For hierarchical cluster analysis take a good look at ?hclust and run its examples. Alternative functions are in the cluster package that comes with R. k-means clustering is available in function kmeans() and also in the cluster package.
A simple hierarchical cluster analysis of the dummy data you show would be done as follows:
## dummy data first
require(MASS)
set.seed(1)
dat <- data.frame(mvrnorm(100, mu = c(2,6,3),
Sigma = matrix(c(10, 2, 4,
2, 3, 0.5,
4, 0.5, 2), ncol = 3)))
Compute the dissimilarity matrix using Euclidean distances (you can use whatever distance you want)
dij <- dist(scale(dat, center = TRUE, scale = TRUE))
Then cluster them, say using the group average hierarchical method
clust <- hclust(dij, method = "average")
Printing the result gives us:
R> clust
Call:
hclust(d = dij, method = "average")
Cluster method : average
Distance : euclidean
Number of objects: 100
Plot the dendrogram
but that simple output belies a complex object that needs further functions to extract or use the information contained therein:
R> str(clust)
List of 7
$ merge : int [1:99, 1:2] -12 -17 -40 -30 -73 -23 1 -52 -91 -45 ...
$ height : num [1:99] 0.0451 0.0807 0.12 0.1233 0.1445 ...
$ order : int [1:100] 84 14 24 67 46 34 49 36 41 52 ...
$ labels : NULL
$ method : chr "average"
$ call : language hclust(d = dij, method = "average")
$ dist.method: chr "euclidean"
- attr(*, "class")= chr "hclust"
The dendrogram can be generated using the plot() method (hang gets the labels at the bottom of the dendrogram, along the x-axis, and cex just shrinks all the labels to 70% or normal)
plot(clust, hang = -0.01, cex = 0.7)
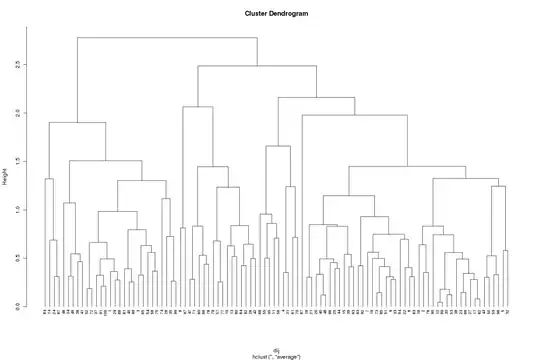
Say we want a 3-cluster solution, cut the dendrogram to produce 3 groups and return the cluster memberships
R> cutree(clust, k = 3)
[1] 1 2 1 2 2 3 2 2 2 3 2 2 3 1 2 2 2 2 2 2 2 2 2 1 2 3 2 1 1 2 2 2 2 1 1 1 1
[38] 2 2 2 1 3 2 2 1 1 3 2 1 2 2 1 2 1 2 2 3 1 2 3 2 2 2 3 1 3 1 2 2 2 3 1 2 1
[75] 1 2 3 3 3 3 1 3 2 1 2 2 2 1 2 2 1 2 2 2 2 2 3 1 1 1
That is, cutree() returns a vector the same length as the number of observations clustered, the elements of which contain the group ID that each observation belongs. The membership is the ID of the leaf into which each observation falls when the dendrogram is cut at a stated height or, as done here, at the appropriate height to provide the stated number of groups.
Perhaps that gives you enough to be going on with?
For k-means, we would do this
set.seed(2) ## *k*-means uses a random start
klust <- kmeans(scale(dat, center = TRUE, scale = TRUE), centers = 3)
klust
which gives
> klust
K-means clustering with 3 clusters of sizes 41, 27, 32
Cluster means:
X1 X2 X3
1 0.04467551 0.69925741 -0.02678733
2 1.11018549 -0.01169576 1.16870206
3 -0.99395950 -0.88605526 -0.95177110
Clustering vector:
[1] 3 1 3 2 2 3 1 1 1 1 2 1 1 3 2 3 1 2 1 2 2 1 1 3 2 1 1 3 3 1 2 2 1 3 3 3 3
[38] 1 2 2 3 1 2 2 3 3 1 2 3 2 1 3 1 3 2 2 1 3 2 1 2 1 1 1 3 1 3 2 1 2 1 3 1 3
[75] 3 1 1 1 1 1 3 1 2 3 1 1 1 3 1 1 3 2 2 1 2 2 3 3 3 3
Within cluster sum of squares by cluster:
[1] 47.27597 31.52213 42.15803
(between_SS / total_SS = 59.3 %)
Available components:
[1] "cluster" "centers" "totss" "withinss" "tot.withinss"
[6] "betweenss" "size"
Here we get some information about the components in the object returned by kmeans(). The $cluster component will yield the membership vector, comparable to the output we saw earlier from cutree():
R> klust$cluster
[1] 3 1 3 2 2 3 1 1 1 1 2 1 1 3 2 3 1 2 1 2 2 1 1 3 2 1 1 3 3 1 2 2 1 3 3 3 3
[38] 1 2 2 3 1 2 2 3 3 1 2 3 2 1 3 1 3 2 2 1 3 2 1 2 1 1 1 3 1 3 2 1 2 1 3 1 3
[75] 3 1 1 1 1 1 3 1 2 3 1 1 1 3 1 1 3 2 2 1 2 2 3 3 3 3
In both instances, notice that I also scale (standardise) the data to allow each variable to be compared on a common scale. With data measured in different "units" or on different scales (as here with different means and variances) this is an important data processing step if the results are to be meaningful or not dominated by the variables that have large variances.
