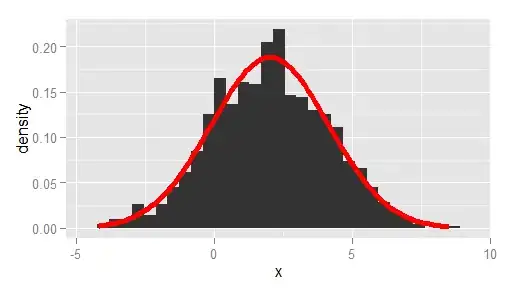
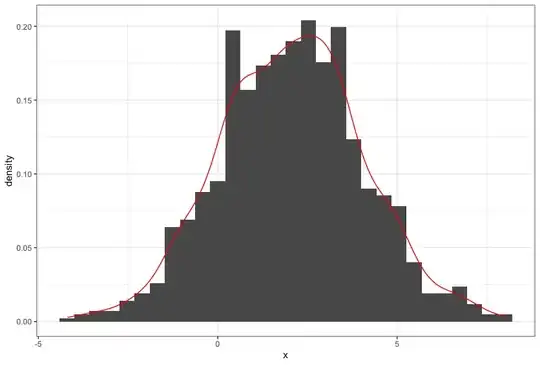
I am trying to make a histogram of density values and overlay that with the curve of a density function (not the density estimate).
Using a simple standard normal example, here is some data:
x <- rnorm(1000)
I can do:
q <- qplot( x, geom="histogram")
q + stat_function( fun = dnorm )
but this gives the scale of the histogram in frequencies and not densities. with ..density.. I can get the proper scale on the histogram:
q <- qplot( x,..density.., geom="histogram")
q
But now this gives an error:
q + stat_function( fun = dnorm )
Is there something I am not seeing?
Another question, is there a way to plot the curve of a function, like curve(), but then not as layer?