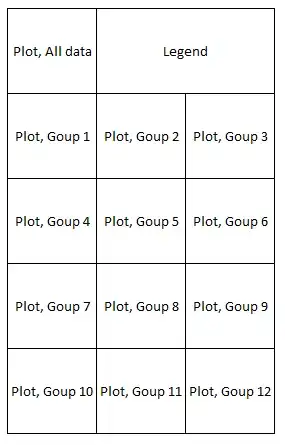
I am trying to plot several plots of "state sequence object" created with the TraMineR package using the plot_grid() function. My main problem is, that I can not store the plots created with the seqplot() function in a list. My problem is not about how to position the legend or "sub"plots but about how to actually plot the seqplots in a grid.
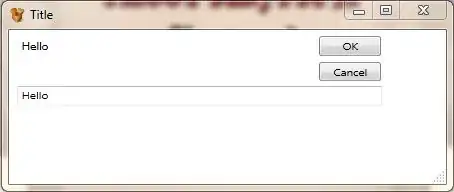
I stored ggscatter plots in a list and passed that list to the plot_grid() function before - so I am relatively sure that this works in general. I think the problem is mainly the object type produced by seqplot(). I tried to store the plots using the as.ggplot() function, which did not work (example A below).
Using the recordPlot() function works more or less. It does not look nice though (example B + C below).
library(TraMineR)
library(ggplotify)
data(biofam)
biofam.lab <- c("Parent", "Left", "Married", "Left+Marr",
"Child", "Left+Child", "Left+Marr+Child", "Divorced")
biofam.seq <- seqdef(biofam[1:600,], 10:25, labels=biofam.lab)
# Example A ---------------------------------------------------------------
# not even able to store the plot
plot.list <- list()
plot.list[[1]] <- as.ggplot(seqplot(biofam.seq, type = "I", with.legend = FALSE, sortv = "from.start"))
# Example B and C ---------------------------------------------------------
plot.list <- list()
seqplot(biofam.seq, type = "I", with.legend = FALSE, sortv = "from.start")
plot.list[[1]] <- recordPlot()
seqplot(biofam.seq, type = "I", with.legend = FALSE, sortv = "from.start")
plot.list[[2]] <- recordPlot()
seqplot(biofam.seq, type = "I", with.legend = FALSE, sortv = "from.start")
plot.list[[3]] <- recordPlot()
seqplot(biofam.seq, type = "I", with.legend = FALSE, sortv = "from.start")
plot.list[[4]] <- recordPlot()
# Example B
plot_grid(plotlist = plot.list, ncol = 2)
# Example C
plot_grid(
plot_grid(plotlist = plot.list, ncol = 2),
plot_grid(plotlist = plot.list, ncol = 2)
)
I want to be relatively free in positioning the actual elements I want to plot in a grid. For example, I want to save a page containing 13 such plots and a legend in a 5 x 3 grid - as can be seen in the example. That's why I think going with plot_grid() works better compared to, e.g., par(mfrow = c(5,3)). Additionally, using par() I get "Error in plot.new() : figure margins too large".