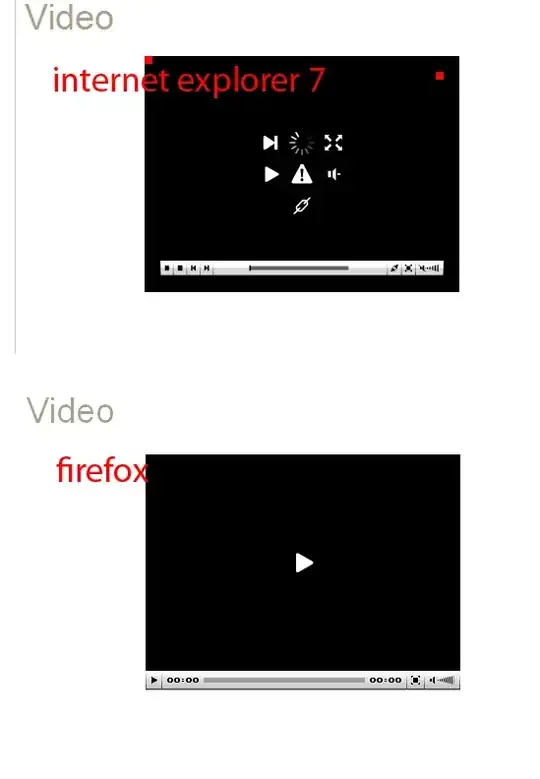
I have 3 dataframes, training_data, validation_data, test_data, and I need to plot them after each other with different colors so that it looks like one line but divided in 3 color. I tried to do that by moving the x-axis start, using xlim, for the second and third time series as following code shows, but it plots all of them startong from x=0. How can I fix it?
train_data.loc[idx].plot(kind='line'
, use_index=False
, color='blue'
, label='Training Data'
, legend=False)
validation_data.loc[idx].plot(kind='line'
, use_index=False
, figsize=(20, 5)
, xlim=362
, color='red'
, label='Validation Data'
, legend=False)
test_data.loc[idx].plot(kind='line'
, use_index=False
, figsize=(20, 5)
, xlim=481
, color='green'
, label='Test Data'
, legend=False)
plt.xlim(xmin=0)
plt.legend(loc=1, prop={'size': 'xx-small'})
plt.savefig("data.pdf")
plt.clf()
plt.close()
UPDATE:
All 3 dataframes has the following shape (N, 28), there are 138 different indexes (idx) and all dataframes have part of each index. Actually, each index is a time series that was splitted to three parts as training, validation and test datasets. I need to plot only the first column, var0, of each index. That's why I'm using <df>.loc[idx].iloc[:, 0]
df=
idx var0 var1 var2 var3 var4 ... var28
5171 10.0 2.8 0.0 5.0 1.0 ... 9.4
5171 40.9 2.5 3.4 4.5 1.3 ... 7.7
5171 60.7 3.1 5.2 6.6 3.4 ... 1.0
...
5171 0.5 1.3 5.1 0.5 0.2 ... 0.4
4567 1.5 2.0 1.0 4.5 0.1 ... 0.4
4567 4.4 2.0 1.3 6.4 0.1 ... 3.3
4567 6.3 3.0 1.5 7.6 1.6 ... 1.6
...
4567 0.7 1.4 1.4 0.3 4.2 ... 1.7
...
9584 0.3 2.6 0.0 5.2 1.6 ... 9.7
9584 0.5 1.2 8.3 3.4 1.3 ... 1.7
9584 0.7 3.0 5.6 6.6 3.0 ... 1.0
...
9584 0.7 1.3 0.1 0.0 2.0 ... 1.7
I tried to combine all three dataframes in one and then plot it using slicing as @Brendan Cox suggested. But I'm not getting the results I need, it still starts the plots from x=0. Here is the code:
data = pd.concat([train_data.loc[idx].iloc[:, 0], validation_data.loc[idx].iloc[:, 0], test_data.loc[idx].iloc[:, 0]])
data.iloc[0:362].plot(kind='line'
, use_index=False
, figsize=(20,5)
, color='blue'
, label='Training Data'
, legend=False)
data.iloc[362:481].plot(kind='line'
, use_index=False
, figsize=(20, 5)
, color='red'
, label='Validation Data'
, legend=False)
data.iloc[481:].plot(kind='line'
, use_index=False
, figsize=(20, 5)
, color='green'
, label='Test Data'
, legend=False)
I attached the resulted plot (which is wrong!). I need to have the red and green lines to continue after the blue line