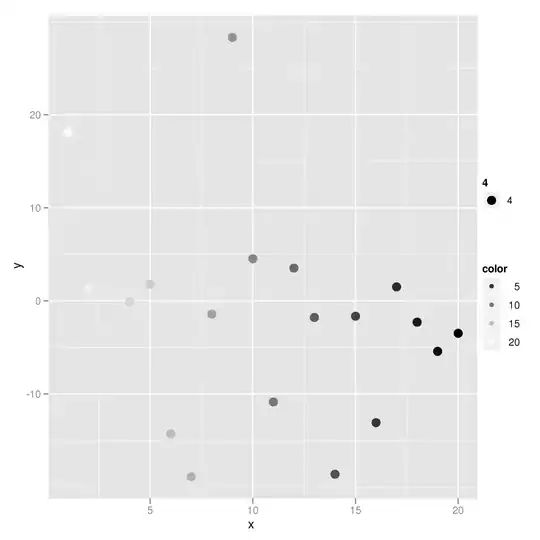
I have a large grid formed by X and Y coordinates, each representing a value. However, some combinations within the grid do not exist, see the attached graphic: 
I would like to identify the missing x-y-combinations with a R script, however don't know how to do this. What's an efficient of getting these combinations?
Example of my data:
df1 <- structure(list(coord_n = c(1065125L, 1065875L, 1064625L, 1064375L,
1065625L, 1065375L, 1065625L, 1065125L, 1065625L, 1065125L, 1066125L,
1064625L, 1066375L, 1064125L, 1064375L, 1064625L, 1066375L, 1064875L,
1066125L, 1066625L, 1064375L, 1065125L, 1066375L, 1066625L, 1065125L,
1065875L, 1064125L, 1064375L, 1064125L, 1065875L, 1064625L, 1065125L,
1065125L, 1065625L, 1066375L, 1064375L, 1064875L, 1065875L, 1066375L,
1066625L, 1064375L, 1064625L, 1066375L, 1065875L, 1065375L, 1065375L,
1066625L, 1065375L, 1064625L, 1066625L, 1066125L, 1065625L, 1065375L,
1065875L, 1064125L, 1064375L, 1064875L, 1065625L, 1065625L, 1064625L,
1064875L, 1065375L, 1065875L, 1065875L, 1066625L, 1065875L, 1064875L,
1066625L, 1064875L, 1064125L, 1066125L, 1064375L, 1066375L, 1064125L,
1066625L, 1065125L, 1064625L, 1065625L, 1066125L, 1064125L, 1066375L,
1066625L, 1066375L, 1064125L, 1064875L, 1065375L, 1064375L, 1065625L,
1065875L, 1065375L, 1066375L, 1064875L, 1064375L, 1066625L, 1064375L,
1065875L, 1064375L, 1065375L, 1064875L, 1066375L), coord_e = c(2418625L,
2419125L, 2421875L, 2418125L, 2421375L, 2422375L, 2421125L, 2418875L,
2418625L, 2420375L, 2419375L, 2420625L, 2418875L, 2420625L, 2419125L,
2420875L, 2419125L, 2419875L, 2418375L, 2421625L, 2422375L, 2422375L,
2422125L, 2422125L, 2420125L, 2421875L, 2421875L, 2420125L, 2422375L,
2420625L, 2419625L, 2418375L, 2419625L, 2418375L, 2419875L, 2420875L,
2421375L, 2422375L, 2422375L, 2418125L, 2418375L, 2419125L, 2418625L,
2418875L, 2419375L, 2421375L, 2421125L, 2419125L, 2418375L, 2419625L,
2418875L, 2420125L, 2419875L, 2420375L, 2420375L, 2419875L, 2420375L,
2422375L, 2421875L, 2422375L, 2419375L, 2420875L, 2421125L, 2421375L,
2419125L, 2419375L, 2421625L, 2418375L, 2418875L, 2418375L, 2420125L,
2419625L, 2418375L, 2420125L, 2421375L, 2422125L, 2419875L, 2420375L,
2420375L, 2418625L, 2421125L, 2420125L, 2421625L, 2419875L, 2419125L,
2420625L, 2418625L, 2419375L, 2420125L, 2418125L, 2420125L, 2418625L,
2418875L, 2418625L, 2421125L, 2419875L, 2421375L, 2418875L, 2420875L,
2421875L), density_value = c(0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L), percentage_free = c(100,
100, 100, 100, 100, 100, 100, 100, 100, 100, 100, 100, 100, 100,
100, 100, 100, 100, 100, 100, 100, 100, 100, 100, 100, 100, 100,
100, 100, 100, 100, 100, 100, 100, 100, 100, 100, 100, 100, 100,
100, 100, 100, 100, 100, 100, 100, 100, 100, 100, 100, 100, 100,
100, 100, 100, 100, 100, 100, 100, 100, 100, 100, 100, 100, 100,
100, 100, 100, 100, 100, 100, 100, 100, 100, 100, 100, 100, 100,
100, 100, 100, 100, 100, 100, 100, 100, 100, 100, 100, 100, 100,
100, 100, 100, 100, 100, 100, 100, 100)), class = c("data.table",
"data.frame"), row.names = c(NA, -100L))