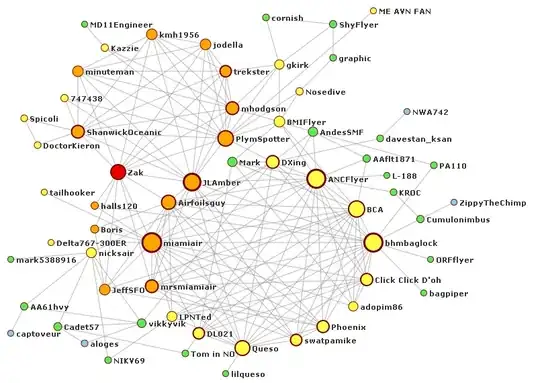
Using igraph in R-studio I made a network analysis based on an adjacency matrix(45*45) that has 45 culture nodes from interviews with multiple people in the same organisations. A connection between 2 nodes was coded "1" whenever 2 cultural themes were spoken about in the same passage, so connections always go both ways and are not directed. The problem with my network analysis is that there are a lot of connections, and the graph is not easy to read, that is why I tried to add in a layout to form some kind of hierarchy so I can see if certain cultural aspects are grouped (and thus if certain culture nodes form a theme together). However, when I add a layout such as my lables in "words" dissapear and I get a smaller amount of nodes with numbers, however I don't know what this numbers mean.
I have tried to use the fruchterman.reingold. layout. However, I get some error saying "l" cannot be found (see code below). Then I get a nice tree like output, however with numbers, which I don't know what they mean. I would like to be able to read what cultural themes are grouped together and thus need to know which ones are closest together/ connect most/ form a group. See the code I used below:
my_data <- read.csv(file.choose(),sep=";",header=TRUE)
nodelist <- names(my_data)[-1]
my_matrix <- as.matrix(my_data) [,-1]
rownames(my_matrix) <- colnames(my_matrix) <- nodelist
my_matrix
library(igraph)
g <- graph_from_adjacency_matrix(my_matrix, mode="undirected",
weighted=NULL)
my_matrix.bg <- barabasi.game(80)
V(my_matrix.bg)$frame.color <- "white"
V(my_matrix.bg)$color <- "orange"
V(my_matrix.bg)$label <- "orange"
V(my_matrix.bg)$size <- 10
E(my_matrix.bg)$arrow.mode <- 0
l <- layout.fruchterman.reingold.
(my_matrix.bg,repulserad=vcount(my_matrix.bg)^3,area=vcount
(my_matrix.bg)^2.4)
par(mfrow=c(1,2), mar=c(0,0,0,0))
plot(my_matrix.bg,layout=layout.fruchterman.reingold)
plot(my_matrix.bg,layout=l)
I hope to get a network analysis output, which is readable in which I can see what nodes have most/ the strongest connections, and can clearly see if nodes exist. It would also be nice If this output can be made larger and readable.