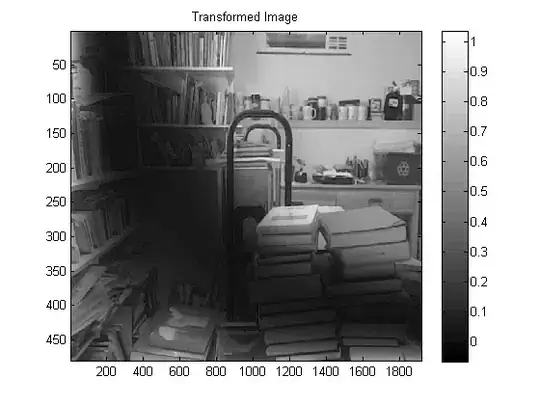
Here's an approach adapted from this blog post
- Convert image to grayscale
- Otsu's threshold to obtain a binary image
- Compute Euclidean Distance Transform
- Perform connected component analysis
- Apply watershed
- Iterate through label values and extract objects
Here's the results
While iterating through each contour, you can accumulate the total area
1388903.5
import cv2
import numpy as np
from skimage.feature import peak_local_max
from skimage.morphology import watershed
from scipy import ndimage
# Load in image, convert to gray scale, and Otsu's threshold
image = cv2.imread('1.jpg')
gray = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
thresh = cv2.threshold(gray, 0, 255, cv2.THRESH_BINARY + cv2.THRESH_OTSU)[1]
# Compute Euclidean distance from every binary pixel
# to the nearest zero pixel then find peaks
distance_map = ndimage.distance_transform_edt(thresh)
local_max = peak_local_max(distance_map, indices=False, min_distance=20, labels=thresh)
# Perform connected component analysis then apply Watershed
markers = ndimage.label(local_max, structure=np.ones((3, 3)))[0]
labels = watershed(-distance_map, markers, mask=thresh)
# Iterate through unique labels
total_area = 0
for label in np.unique(labels):
if label == 0:
continue
# Create a mask
mask = np.zeros(gray.shape, dtype="uint8")
mask[labels == label] = 255
# Find contours and determine contour area
cnts = cv2.findContours(mask.copy(), cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE)
cnts = cnts[0] if len(cnts) == 2 else cnts[1]
c = max(cnts, key=cv2.contourArea)
area = cv2.contourArea(c)
total_area += area
cv2.drawContours(image, [c], -1, (36,255,12), 4)
print(total_area)
cv2.imshow('image', image)
cv2.waitKey()