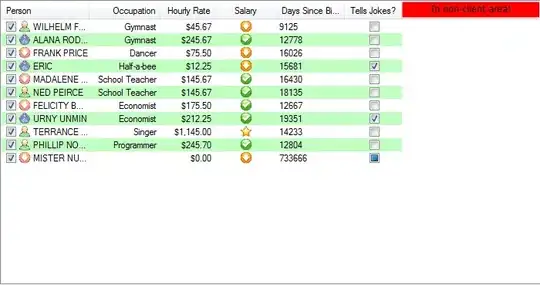
I'm trying to create a 2D line chart with seaborn, but I get several artefacts as seen here, i.e. lines that suddenly shoot down or up with barely-visible vertical lines:
Excel on the other hand produces a correct visualisation from the same file:
My code follows the seaborn examples (a sample test.csv can be found here):
import seaborn as sns
import pandas as pd
import matplotlib.pyplot as plt
data = pd.read_csv('test.csv')
sns.set()
lp = sns.lineplot(x=data['x'], y=data['y'], sort=False, lw=1)
plt.show()
Am I doing something wrong, or is matplotlib unable to handle overlapping values?