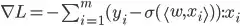Note that this algorithm is memory bandwidth limited. If you optimize this in a larger context (real application) there is very likely a higher speedup possible.
Example
import numpy as np
#https://stackoverflow.com/a/29863846/4045774
def sigmoid(x):
return np.exp(-np.logaddexp(0, -x))
def calc_loss_grad_1(weights, X_batch, y_batch):
n_samples, n_features = X_batch.shape
loss_grad = np.zeros((n_features,))
for i in range(n_samples):
sigm = sigmoid(X_batch[i,:] @ weights)
loss_grad += - (y_batch[i] - sigm) * X_batch[i]
return loss_grad
def calc_loss_grad_2(weights, X_batch, y_batch):
sigm =-y_batch+sigmoid(X_batch@weights)
return sigm@X_batch
weights=np.random.rand(n_features)
X_batch=np.random.rand(n_samples,n_features)
y_batch=np.random.rand(n_samples)
#n_samples=int(1e5)
#n_features=int(1e4)
%timeit res=calc_loss_grad_1(weights, X_batch, y_batch)
#1.79 s ± 35.8 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
%timeit res=calc_loss_grad_2(weights, X_batch, y_batch)
#539 ms ± 21.9 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
#n_samples=int(1e3)
#n_features=int(1e2)
%timeit res=calc_loss_grad_1(weights, X_batch, y_batch)
#3.68 ms ± 44.7 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
%timeit res=calc_loss_grad_2(weights, X_batch, y_batch)
#49.1 µs ± 503 ns per loop (mean ± std. dev. of 7 runs, 10000 loops each)