I'm posting because I am numerically solving ordinary differential equations and would like to constrain the state variables. In sum, I'm a biologist modeling populations and would like for the populations to go extinct when they become really small.
When I run my models populations can become very, very small (e.g., 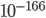 ), but never go to 0. It's important for me that they do so, so I can calculate extinction. But more realistically, when a population is at a density that is considerably smaller than there are at atoms on Earth, that's not too realistic ;)
), but never go to 0. It's important for me that they do so, so I can calculate extinction. But more realistically, when a population is at a density that is considerably smaller than there are at atoms on Earth, that's not too realistic ;)
Even in the simple 2-species enemy-victim model like the following reaches densities of 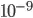 that I would like to be interpreted as 0.
that I would like to be interpreted as 0.
library(package = "deSolve")
lv <- function(times, state, parms) {
with(as.list(c(state, parms)), {
dR <- 2*R - 0.5*R*C
dC <- 0.2*R*C - 0.6*C
return(list(c(dR, dC)))
})
}
time_vec <- seq(from = 0, to = 100, length.out = 1e4)
y_0 <- c(R = 50, C = 20)
out <- ode(y = y_0, times = time_vec, func = lv, parms = NULL, method = "lsoda")
min(out[,-1])
plot(x = out[,2], y = out[,3], type = "l")
I'd ideally like to see how extinction can be modeled using the solver `deSolve' in R, but any help directing me towards general answers/names for this type of problem would also be very appreciated.
P.S. This is similar to a post on wanting to replace negative values with 0 (link), but different because I want the populations not to just be non-negative, but stay at 0 infinitely. But also, there wasn't a good answer on how to do that in this post.