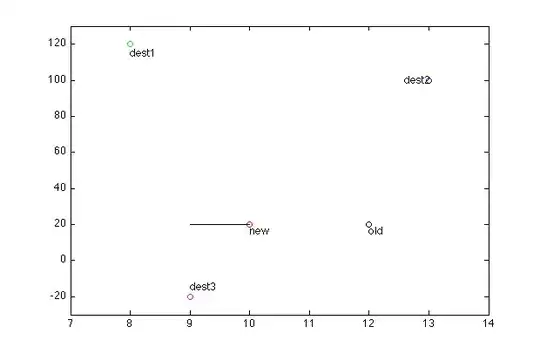
I have a table that looks like this:
code to reproduce this dataframe in R:
ID = c("0", "51", "7", "62", "1","10","5", "79", "62", "10","1","7")
mRNA = c("0", "0", "30", "1", "0", "14", "0", "1", "1", "16", "0", "0")
Centroid = c("d0","d0", "d0", "d0", "d1", "d1","d1", "d1", "d1", "d10", "d10", "d10")
df <- data.frame(ID,mRNA,Centroid)
I would like to reformat this data to look like this:
So far I have tried this:
r <- reshape(df, direction = "wide", idvar="Centroid", timevar="ID")
The output looks close to what I want but not there yet (I don't want it to rename the columns using the IDs).
I prefer solutions in R, but if you have suggestions in python I can also try them out. Any help would be very much appreciated!