If you are willing to start from a NumPy array, you could adapt the solutions from here, omitting the senseless ones like those using zip() or the key parameter from min()/max()), and adding a couple more:
def extrema_py(arr):
return max(arr[:, 0]), min(arr[:, 0]), max(arr[:, 1]), min(arr[:, 1])
import numpy as np
def extrema_np(arr):
return np.max(arr[:, 0]), np.min(arr[:, 0]), np.max(arr[:, 1]), np.min(arr[:, 1])
import numpy as np
def extrema_npt(arr):
arr = arr.transpose()
return np.max(arr[0, :]), np.min(arr[0, :]), np.max(arr[1, :]), np.min(arr[1, :])
import numpy as np
def extrema_npa(arr):
x_max, y_max = np.max(arr, axis=0)
x_min, y_min = np.min(arr, axis=0)
return x_max, x_min, y_max, y_min
import numpy as np
def extrema_npat(arr):
arr = arr.transpose()
x_max, y_max = np.max(arr, axis=1)
x_min, y_min = np.min(arr, axis=1)
return x_max, x_min, y_max, y_min
def extrema_loop(arr):
n, m = arr.shape
x_min = x_max = arr[0, 0]
y_min = y_max = arr[0, 1]
for i in range(1, n):
x, y = arr[i, :]
if x > x_max:
x_max = x
elif x < x_min:
x_min = x
if y > y_max:
y_max = y
elif y < y_min:
y_min = y
return x_max, x_min, y_max, y_min
import numba as nb
@nb.jit(nopython=True)
def extrema_loop_nb(arr):
n, m = arr.shape
x_min = x_max = arr[0, 0]
y_min = y_max = arr[0, 1]
for i in range(1, n):
x = arr[i, 0]
y = arr[i, 1]
if x > x_max:
x_max = x
elif x < x_min:
x_min = x
if y > y_max:
y_max = y
elif y < y_min:
y_min = y
return x_max, x_min, y_max, y_min
%%cython -c-O3 -c-march=native -a
#cython: language_level=3, boundscheck=False, wraparound=False, initializedcheck=False, cdivision=True, infer_types=True
import numpy as np
import cython as cy
cdef void _extrema_loop_cy(
long[:, :] arr,
size_t n,
size_t m,
long[:, :] result):
cdef size_t i, j
cdef long x, y, x_max, x_min, y_max, y_min
x_min = x_max = arr[0, 0]
y_min = y_max = arr[0, 1]
for i in range(1, n):
x = arr[i, 0]
y = arr[i, 1]
if x > x_max:
x_max = x
elif x < x_min:
x_min = x
if y > y_max:
y_max = y
elif y < y_min:
y_min = y
result[0, 0] = x_max
result[0, 1] = x_min
result[1, 0] = y_max
result[1, 1] = y_min
def extrema_loop_cy(arr):
n, m = arr.shape
result = np.zeros((2, m), dtype=arr.dtype)
_extrema_loop_cy(arr, n, m, result)
return result[0, 0], result[0, 1], result[1, 0], result[1, 1]
and their respective timings as a function of input size:
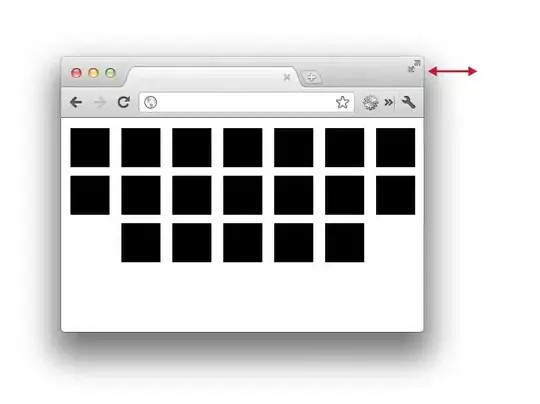

So, for NumPy array inputs, one can gets much faster timings.
The Numba- and Cython- based solution seems to be the fastest, remarkably outperforming the fastest NumPy-only approaches.
(full benchmarks available here)
(EDITED to improve Numba-based solution)