I'd like to plot several data into a grid using holoviews/hvplot, based on one dimension, which contains several unique data points.
Considering this example:
import seaborn as sns
import hvplot.pandas
iris = sns.load_dataset('iris')
plot = iris.hvplot.scatter(x="sepal_length", y="sepal_width", col="species")
hvplot.show(plot)
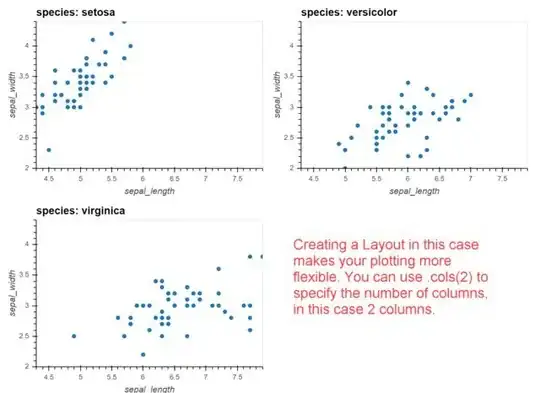
The above code creates several plots, based on the species part of the iris data set, resulting in the picture below:
But now imagine there were not 3 different species, but twenty. The plot would get to wide so I'd like to break the line after a few plots. But I couldn't find any "maximum columns" parameter. A normal grid expects another column to define the rows which I don't have.
Any suggestions would help.