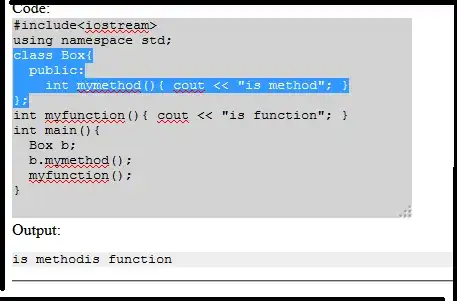
Here is an example and the output is a png file hsa04110.gse16873.png. My question is how to get the plot displayed directly rather than save it as a file.
library(pathview)
data(gse16873.d)
data(demo.paths)
data(paths.hsa)
pathview(gene.data = gse16873.d[, 1], pathway.id = demo.paths$sel.paths[1],
species = "hsa", out.suffix = "gse16873", kegg.native = T)
#> 'select()' returned 1:1 mapping between keys and columns
#> Info: Working in directory D:/github/RNASeq-KEGG
#> Info: Writing image file hsa04110.gse16873.png