I have a pretty complicated case at hand with ggplot2. I tried to exemplify it with a MWE using iris data below.
I just have boxplots in facets, and wanted to move the legend to take the space of the empty facets.
This is all good, I use lemon::reposition_legend() for that and it works.
However, I then have to modify a bunch of things in the plot (namely add significant test results and other things that are not relevant for this question), and I am forced to use ggplot_build() on my output plot for that purpose.
After using ggplot_build() to modify my plot, I do not seem to be able to use reposition_legend() successfully anymore...
Check out my MWE below.
First I load the packages I need, and define a shift_legend() function (which uses reposition_legend()), based on an answer to this question.
library(tidyr)
library(ggplot2)
library(ggplotify)
library(gtable)
library(cowplot)
library(purrr)
library(lemon)
library(grid)
shift_legend <- function(p) {
pnls <- NULL
if (class(p)[1] == "gtable") pnls <- p
else if (class(p)[2] == "ggplot") pnls <- plot_to_gtable(p)
else stop("Please provide a ggplot or a gtable object")
pnls <- gtable_filter(pnls, "panel")
pnls <- setNames(pnls$grobs, pnls$layout$name)
pnls <- keep(pnls, ~identical(.x, zeroGrob()))
res <- NULL
if(length(pnls) > 0) res <- reposition_legend( p, "center", panel=names(pnls) )
else res <- p
return(res)
}
I then load the iris data and make my plot with shift_legend() successfully.
data(iris)
summary(iris)
iris_long <- gather(iris, "Variable", "Value", -Species)
P <- ggplot(iris_long, aes(x=Variable, y=Value)) +
geom_boxplot(aes(fill=Variable), position=position_dodge(.9)) +
facet_wrap(.~Species, ncol=2) +
theme_light() +
theme(legend.key.size = unit(0.5, "inch"))
out_file_name <- "test.pdf"
pdf(file=out_file_name, height=10, width=10, onefile=FALSE)
print(
grid.draw(shift_legend(P))
)
dev.off()
This produces this output, all good till here:
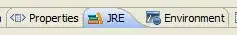 Note this is the arrangement I want to be able to reproduce (after using
Note this is the arrangement I want to be able to reproduce (after using ggplot_build), with the legend taking the empty facets space.
But now I need to use ggplot_build() to add and modify things in my plot. After that I can plot it normally without using reposition_legend().
P2 <- ggplot_build(P)
#Do a bunch of things here...
out_file_name2 <- "test2.pdf"
pdf.options(reset=TRUE, onefile=FALSE)
pdf(file=out_file_name2, height=10, width=10)
print(
plot(ggplot_gtable(P2))
)
dev.off()
But I still want to reposition the legend, so I attempt to use reposition_legend() again converting the ggplot_built object into a gtable object (which, according to the function documentation it can accept also as input).
out_file_name22 <- "test22.pdf"
pdf.options(reset=TRUE, onefile=FALSE)
pdf(file=out_file_name22, height=10, width=10)
print(
grid.draw(shift_legend(
ggplot_gtable(P2)
))
)
dev.off()
Here I get this error:
Error in reposition_legend(p, "center", panel = names(pnls)) : No legend given in arguments, or could not extract legend from plot.
I tried again converting the gtable object into a ggplot one using ggplotify::as.ggplot(). This time I obtained no errors, but the legend was not repositioned as expected...
out_file_name222 <- "test222.pdf"
pdf.options(reset=TRUE, onefile=FALSE)
pdf(file=out_file_name222, height=10, width=10)
print(
grid.draw(shift_legend(
as.ggplot(ggplot_gtable(P2))
))
)
dev.off()
Help please!
EDIT
I tried to change the workflow, as suggested in the comments and answers, to no avail.
Being P the original plot, what I need to modify is in the ggplot_build(P)$data data frame.
This data frame looks like this:
> ggplot_build(P)$data
[[1]]
fill ymin lower middle upper ymax outliers notchupper notchlower x PANEL group ymin_final ymax_final xmin xmax weight colour size alpha shape
1 #F8766D 1.2 1.400 1.50 1.575 1.7 1.1, 1.0, 1.9, 1.9 1.5391030 1.4608970 1 1 1 1.0 1.9 0.625 1.375 1 grey20 0.5 NA 19
2 #7CAE00 0.1 0.200 0.20 0.300 0.4 0.5, 0.6 0.2223446 0.1776554 2 1 2 0.1 0.6 1.625 2.375 1 grey20 0.5 NA 19
3 #00BFC4 4.3 4.800 5.00 5.200 5.8 5.0893783 4.9106217 3 1 3 4.3 5.8 2.625 3.375 1 grey20 0.5 NA 19
4 #C77CFF 2.9 3.200 3.40 3.675 4.2 4.4, 2.3 3.5061367 3.2938633 4 1 4 2.3 4.4 3.625 4.375 1 grey20 0.5 NA 19
5 #F8766D 3.3 4.000 4.35 4.600 5.1 3 4.4840674 4.2159326 1 2 1 3.0 5.1 0.625 1.375 1 grey20 0.5 NA 19
6 #7CAE00 1.0 1.200 1.30 1.500 1.8 1.3670337 1.2329663 2 2 2 1.0 1.8 1.625 2.375 1 grey20 0.5 NA 19
7 #00BFC4 4.9 5.600 5.90 6.300 7.0 6.0564120 5.7435880 3 2 3 4.9 7.0 2.625 3.375 1 grey20 0.5 NA 19
8 #C77CFF 2.0 2.525 2.80 3.000 3.4 2.9061367 2.6938633 4 2 4 2.0 3.4 3.625 4.375 1 grey20 0.5 NA 19
9 #F8766D 4.5 5.100 5.55 5.875 6.9 5.7231705 5.3768295 1 3 1 4.5 6.9 0.625 1.375 1 grey20 0.5 NA 19
10 #7CAE00 1.4 1.800 2.00 2.300 2.5 2.1117229 1.8882771 2 3 2 1.4 2.5 1.625 2.375 1 grey20 0.5 NA 19
11 #00BFC4 5.6 6.225 6.50 6.900 7.9 4.9 6.6508259 6.3491741 3 3 3 4.9 7.9 2.625 3.375 1 grey20 0.5 NA 19
12 #C77CFF 2.5 2.800 3.00 3.175 3.6 3.8, 2.2, 3.8 3.0837922 2.9162078 4 3 4 2.2 3.8 3.625 4.375 1 grey20 0.5 NA 19
linetype
1 solid
2 solid
3 solid
4 solid
5 solid
6 solid
7 solid
8 solid
9 solid
10 solid
11 solid
12 solid
I modify aspects of it like annotation (not applicable in this MWE) and colour.
However, if, as suggested, I attempt to shift the legend of P before using ggplot_build() to extract and modify the relevant info, I have to do the following:
P2 <- as.ggplot(shift_legend(P))
ggplot_build(P2)$data
The first command opens a new plotting window, which is undesired.
The second command produces this:
> ggplot_build(P2)$data
[[1]]
x y PANEL group
1 0 0 1 -1
2 1 1 1 -1
[[2]]
PANEL group xmin xmax ymin ymax
1 1 -1 0 1 0 1
This looks nothing like the data data frame I modify in P... Any clue where to find it, if possible, in P2 now?
EDIT 2
Just so you see an example of my real life boxplots to see why modifying ggplot_build(P)$data is important to me.
There is no way to show only significant pairwise comparisons with geom_signif().
What I do is use geom_signif() with dummy text to populate the annotation data frame I can access at ggplot_build(P)$data[[3]], and then add my actual significance values to the $annotation column, and subset the data frame accordingly to show only significant comparisons. There I have full control, and can change the colors of the comparisons according to significance, which group has a higher mean, etc, etc.
I asked this a while ago here, and ever since I have polished this and wrapped it into a function.
As you see, this clashes with my shift_legend function, as I do not seem to find a way to access the data data frame...
This is what I have so far with my real life data, I placed the legend at the bottom, but it would be optimal that it took the empty facets space, especially cause I have cases where there are more empty facets.