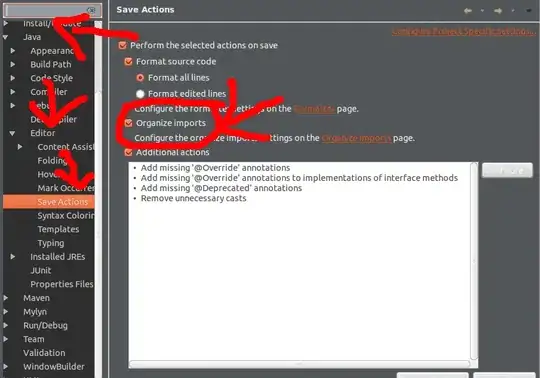
I'm trying to get used to using datetime data in Pandas and plotting different comparisons for a given dataset. I'm using the London Air Quality dataset for Ozone to practice and am trying to replicate the chart below (that I've created using a pivot table in Excel) with Pandas and matplotlib.
The chart plots an average of each hours Ozone reading for each location across the entire dataset to see if there is one location which is constantly higher than others or if different locations have the highest Ozone levels at different periods throughout the day.
Essentially, I'm looking to plot the hourly average of Ozone for each location.
I've attempted to reshape the data into a multiindex format and then plot, similar to what I'd do in excel before plotting but am unsure if this is the correct way to approach the problem. Code for reshaping is below. I am still getting used to reshaping so not sure if this is the correct use/I am approaching the problem in the correct way and open to other methods to accomplish this task. Any assistance to accomplish this task would be much appreciated!
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import datetime
data = pd.read_csv('/Users/xx/Downloads/LaqnData.csv')
data['ReadingDateTime'] = pd.to_datetime(data['ReadingDateTime'])
data['Date'] = pd.to_datetime(data['ReadingDateTime']).dt.date
data['Time'] = pd.to_datetime(data['ReadingDateTime']).dt.time
data.set_index(['Date', 'Time'], inplace = True)
hourly_dataframe = data.pivot_table(columns = 'Site', values = 'Value', index = ['Date', 'Time'])
hourly_dataframe.fillna(method = 'ffill', inplace = True)
hourly_dataframe[hourly_dataframe < 0] = 0