I am using the package growthcurver to create a graph with a sigmoidal curve. It needs to have a logarithmic scale on the y axis.
This works to create a graph without a log scale:
# install.packages("growthcurver")
library("growthcurver")
gcfit <- SummarizeGrowth(curveA$time, curveA$biomass)
gcfit
plot(gcfit)
I have tried plot(gcfit, log=y) and plot(gcfit, log="curveA$biomass"). This gives me the error
'Non-numeric argument to mathematical function'.
Could it be that I am using a data frame? How do I get around this?
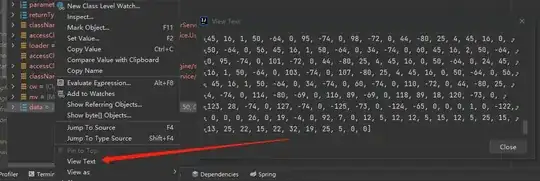
dput(curveA)
structure(list(time = c(1, 2, 3, 4, 5, 6, 7, 8, 17, 18, 19, 20,
21, 22, 23, 24, 25), biomass = c(0.153333333, 1.303333333, 2.836666667,
4.6, 6.21, 6.746666667, 7.283333333, 7.973333333, 8.663333333,
9.046666667, 10.19666667, 10.50333333, 11.04, 11.88333333, 11.96,
11.96, 9.966666667)), class = c("spec_tbl_df", "tbl_df", "tbl",
"data.frame"), row.names = c(NA, -17L), spec = structure(list(
cols = list(time = structure(list(), class = c("collector_number",
"collector")), biomass = structure(list(), class = c("collector_number",
"collector"))), default = structure(list(), class = c("collector_guess",
"collector")), skip = 1), class = "col_spec"))