I'm running a Pyspark process that works without issuess. The first step of the process is to apply specific UDF to the dataframe. This is the function:
import html2text
class Udfs(object):
def __init__(self):
self.h2t = html2text.HTML2Text()
self.h2t.ignore_links = True
self.h2t.ignore_images = True
def extract_text(self, raw_text):
try:
texto = self.h2t.handle(raw_text)
except:
texto = "PARSE HTML ERROR"
return texto
Here is how I apply the UDF:
import pyspark.sql.functions as f
import pyspark.sql.types as t
from udfs import Udfs
udfs = Udfs()
extract_text_udf = f.udf(udfs.extract_text, t.StringType())
df = df.withColumn("texto", extract_text_udf("html_raw"))
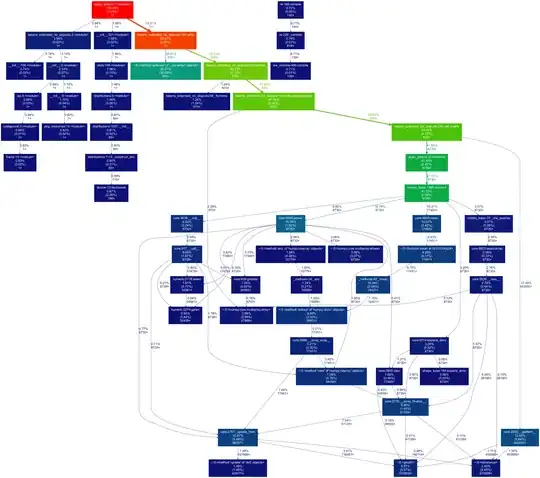
It process approximately 29 million rows and 300GB. The problem is that some tasks takes too much time to process. The average times of the tasks are:
Other tasks have finished with a duration higher than 1 hour.
But some tasks takes too much time processing:
The process run in AWS with EMR in a cluster with 100 nodes, each node with 32gb of RAM and 4 CPUs. Also spark speculation is enabled.
Where is the problem with these tasks? It's a problem with the UDF? It's a thread problem?