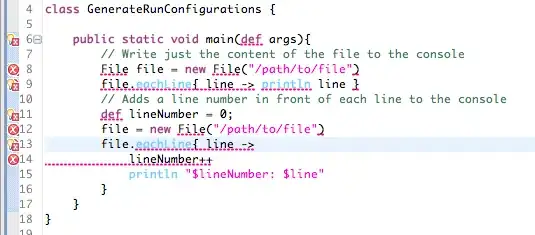
First of all, I would like to say sorry for your confusion to my description because of my poor English. I try to explain my question as my best, if you have anything that not understand please add a comment, I will explain with more details.
The data set used to draw plot like that (the image here is just a part of):
I have put a output of the dput at below.
That is a movement data captured by linear accelerator with the timestamp. I use ggplot2 to draw a line plot to show that in my report. There is my code:
......
#Convert timestamp format
time <- gsub(":", ".", x)
time <- strptime(time, format = "%H.%M.%OS")
time <- as.POSIXct(time)
df["time"] <- time
# Person B Plot
p <- ggplot(df, aes(x = time)) +
scale_x_datetime(name = "Time", labels = date_format("%H:%M:%OS")) +
ylab("PCA") +
geom_hline(aes(yintercept = 0)) +
scale_colour_manual("", values = c("PCA_A" = "hotpink3", "PCA_B" = "steelblue3", "Correlation" = "chocolate")) +
geom_line(aes(y = PCA_b, group = 1, colour = "PCA_B"), size = 0) +
# theme(text = element_text(size = 23), plot.title = element_text(hjust = 0.5)) +
ggtitle("PCA_Two")
Because of the timestamp stored in the csv file as string. I have to change the format to POSIXct, then I can use scale_x_datetime to show the time on the x axis.Then I get a strange plot.
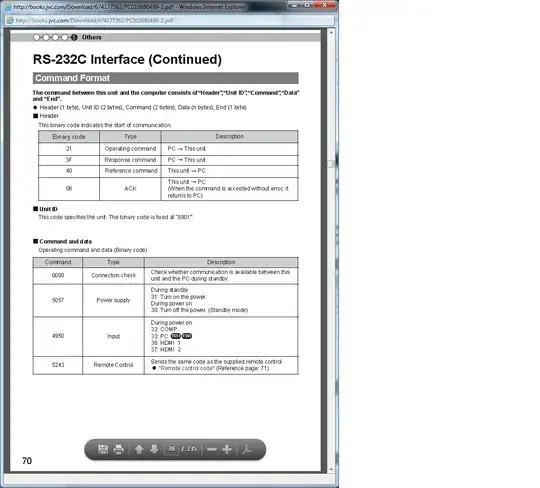
There is a break between the two point. If I remove the first five lines and the "scale_x_datetime" in the code I showed. The plot will be fine, the curve will be smooth but the x axis can not show the time correctly.
Why and How?
---------- update 20/4/2020
I use the dput(df[20:50,]) to output a part of my dataset, I hope that will be helpful. Thanks the help from @chemdork123.
There is a simple description for the data structure below. The dataframe used to draw plot have four columns, time, PCA_a, PCA_b, cor. I will draw three line plot, all the three plot's X data is time (timestamp). In this post, I just show the "time - PCA_b" plot. In fact, all the three plot have the same issue, the break, and the break locations are same. (The NA in the "cor" col is not a bug, that's what I did on purpose.)
structure(list(time = structure(c(1587503540.556, 1587503540.577,
1587503540.615, 1587503540.637, 1587503540.675, 1587503540.696,
1587503540.716, 1587503540.756, 1587503540.776, 1587503540.817,
1587503540.837, 1587503540.876, 1587503540.893, 1587503540.915,
1587503540.937, 1587503540.976, 1587503540.997, 1587503541.018,
1587503541.059, 1587503541.078, 1587503541.117, 1587503541.138,
1587503541.18, 1587503541.201, 1587503541.24, 1587503541.26,
1587503541.3, 1587503541.339, 1587503541.358, 1587503541.4, 1587503541.423
), class = c("POSIXct", "POSIXt"), tzone = ""), PCA_a = c(1.56737319252217,
2.04606254627585, 2.49366222484302, 2.88101522283612, 3.18379411504211,
3.38503090762478, 3.47436865063648, 3.44747654856326, 3.30707775976109,
3.06371801441373, 2.73437161733756, 2.33935677190782, 1.89968708587307,
1.43586301558354, 0.967277030171067, 0.511214148600076, 0.0816220889456876,
-0.311381715806983, -0.661355048674678, -0.965683235694069, -1.22624198074107,
-1.44997061419577, -1.64740413737597, -1.82782646420492, -1.99421995781177,
-2.14199256386341, -2.26073408401317, -2.33585157388011, -2.34937651266747,
-2.28185734041769, -2.11603996134387), PCA_b = c(0.428589019048672,
0.437715207869297, 0.44415836273225, 0.447676595545035, 0.448336071890988,
0.446396459498192, 0.442205853553038, 0.43616876635858, 0.42877854629294,
0.420603253124693, 0.412148862183822, 0.403676755189904, 0.395124979959946,
0.386241966203463, 0.376849622459395, 0.367015680942488, 0.35712348581213,
0.347977244142877, 0.340825041944267, 0.337103574812562, 0.338073413214583,
0.344591707232845, 0.35695103029739, 0.374713701538921, 0.396660690638421,
0.420888192551911, 0.445042523797771, 0.466693774961235, 0.483678597255532,
0.494312865435414, 0.497599592736315), cor = c(0.787242026266416,
NA, NA, NA, NA, NA, NA, NA, NA, 0.297936210131332, NA, NA, NA,
NA, NA, NA, NA, NA, -0.074108818011257, NA, NA, NA, NA, NA, NA,
NA, NA, -0.437523452157598, NA, NA, NA)), row.names = 20:50, class = "data.frame")
---------- update 21/4/2020
I found a very interesting thing. If the size of the dataset smaller than 277, the plot will be perfect. Or the No.277 point will shift. I make a gist here with a 277 size dput. Anyone can test it? My plot will be