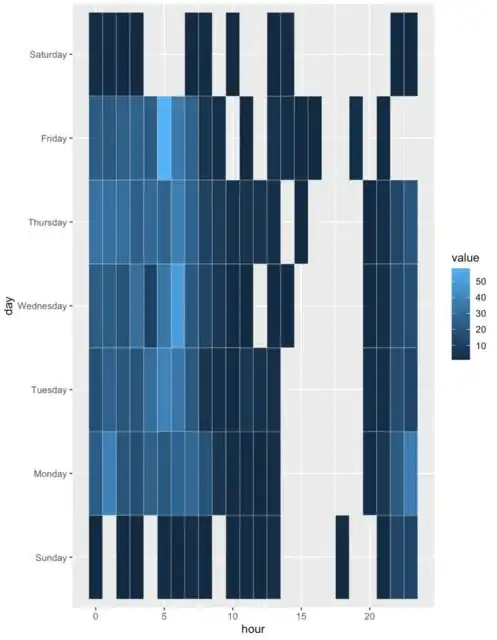
Here is a geom_tile displaying hours and days of the week, how can it made to display each hour (i.e. 00:00 through to 23:00 on the x axis)?
library(tidyverse)
df %>%
ggplot(aes(hour, day, fill = value)) +
geom_tile(colour = "ivory")
Currently it displays every fifth hour:
I have tried a bunch of different things, and would prefer a 'best practice' way (i.e. without manually generating labels), but in case labels are needed, here's one way to produce them hour_labs <- 0:23 %>% { ifelse(nchar(.) == 1, paste0("0", .), .) } %>% paste0(., ":00")
Data for reproducible example
df <- structure(list(day = structure(c(1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L,
4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 5L, 5L,
5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L,
5L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L,
6L, 6L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L), .Label = c("Sunday",
"Monday", "Tuesday", "Wednesday", "Thursday", "Friday", "Saturday"
), class = c("ordered", "factor")), hour = c(0L, 2L, 3L, 5L,
6L, 7L, 8L, 10L, 11L, 12L, 13L, 18L, 21L, 22L, 23L, 0L, 1L, 2L,
3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L, 13L, 20L, 21L, 22L,
23L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L, 13L,
20L, 21L, 22L, 23L, 0L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L,
11L, 13L, 14L, 20L, 21L, 22L, 23L, 0L, 1L, 2L, 3L, 4L, 5L, 6L,
7L, 8L, 9L, 10L, 11L, 12L, 13L, 15L, 20L, 21L, 22L, 23L, 0L,
1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 11L, 13L, 14L, 15L, 16L,
19L, 21L, 0L, 1L, 2L, 3L, 7L, 8L, 10L, 13L, 14L, 22L, 23L), value = c(1L,
1L, 1L, 2L, 1L, 3L, 1L, 1L, 2L, 1L, 3L, 1L, 2L, 13L, 13L, 24L,
39L, 21L, 17L, 25L, 22L, 27L, 28L, 19L, 6L, 2L, 2L, 1L, 2L, 2L,
7L, 23L, 38L, 18L, 26L, 21L, 20L, 31L, 40L, 35L, 22L, 5L, 3L,
2L, 7L, 4L, 3L, 3L, 3L, 17L, 13L, 23L, 24L, 19L, 31L, 13L, 35L,
50L, 22L, 13L, 7L, 2L, 1L, 1L, 1L, 1L, 3L, 14L, 17L, 33L, 32L,
32L, 25L, 29L, 27L, 38L, 26L, 11L, 8L, 4L, 5L, 5L, 3L, 1L, 1L,
3L, 14L, 21L, 24L, 22L, 25L, 26L, 23L, 58L, 36L, 26L, 6L, 3L,
1L, 5L, 3L, 1L, 1L, 3L, 1L, 2L, 2L, 1L, 1L, 1L, 2L, 1L, 1L, 2L,
1L, 1L)), row.names = c(NA, -116L), groups = structure(list(day = structure(1:7, .Label = c("Sunday",
"Monday", "Tuesday", "Wednesday", "Thursday", "Friday", "Saturday"
), class = c("ordered", "factor")), .rows = structure(list(1:15,
16:33, 34:51, 52:69, 70:88, 89:105, 106:116), ptype = integer(0), class = c("vctrs_list_of",
"vctrs_vctr"))), row.names = c(NA, 7L), class = c("tbl_df", "tbl",
"data.frame"), .drop = TRUE), class = c("grouped_df", "tbl_df",
"tbl", "data.frame"))