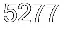I would like to sample points from a normal distribution, and then build up a dotplot one by one using the gganimate package until the final frame shows the full dotplot.
A solution that works for larger datasets ~5,000 - 20,000 points is essential.
Here is the code I have so far:
library(gganimate)
library(tidyverse)
# Generate 100 normal data points, along an index for each sample
samples <- rnorm(100)
index <- seq(1:length(samples))
# Put data into a data frame
df <- tibble(value=samples, index=index)
The df looks like this:
> head(df)
# A tibble: 6 x 2
value index
<dbl> <int>
1 0.0818 1
2 -0.311 2
3 -0.966 3
4 -0.615 4
5 0.388 5
6 -1.66 6
The static plot shows the correct dotplot:
# Create static version
plot <- ggplot(data=df, mapping=aes(x=value))+
geom_dotplot()
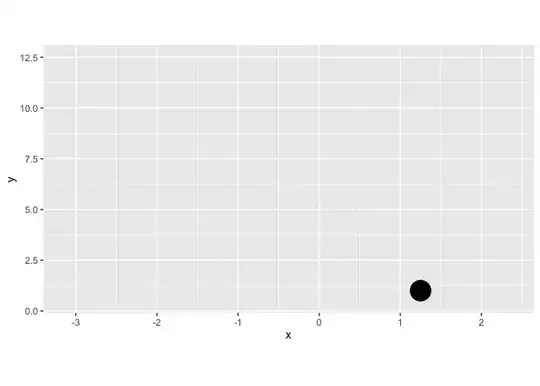
However, the gganimate version does not (see below). It only puts the dots on the x-axis and doesn't stack them.
plot+
transition_reveal(along=index)
Something similar to this would be ideal:
Credit: https://gist.github.com/thomasp85/88d6e7883883315314f341d2207122a1