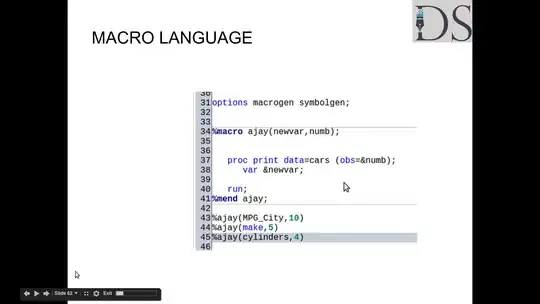
I have a bimodal distribution for the range [-0.1, 0.1] which can be viewed here:
I want to train/fit a Kernel Density Estimation (KDE) on the bimodal distribution as shown in the picture and then, given any other distribution say a uniform distribution such as:
# a uniform distribution between the same range [-0.1, 0.1]-
u_data = np.random.uniform(low = -0.1, high = 0.1, size = (1782,))
I want to be able to use the trained KDE to 'predict' how many of the data points from the given data distribution (say, 'u_data') belong to the target bimodal distribution.
I tried the following code but it doesn't work out:
# Here 'a' is the numpy array containing target bimodal distribution.
# Generate random samples-
kde_samples = {}
for kernel in ['tophat', 'gaussian']:
# Train a kernel on bimodal data distribution 'a'-
kde = KernelDensity(kernel=kernel, bandwidth=0.2).fit(a.reshape(-1, 1))
# Try and generate 300 random samples from trained model-
kde_samples[kernel] = np.exp(kde.sample(300))
# Visualize data distribution using histograms-
plt.hist(a, bins=20, label = 'original distribution')
# sns.distplot(a, kde = True, bins = 20, label = 'original distribution')
plt.hist(kde_samples['gaussian'], bins = 20, label = 'KDE: Gaussian')
plt.hist(kde_samples['tophat'], bins = 20, label = 'KDE: tophat')
plt.title("KDE: Data distribution")
plt.xlabel("weights")
plt.ylabel("frequency")
plt.legend(loc = 'best')
plt.show()
This gives the following visualization:
Two things are wrong:
- The range of the generated samples are wrong!
- The distribution of generated data is NOT bimodal
How can I therefore: train/fit a Kernel Density Estimation (KDE) on the bimodal distribution and then, given any other distribution (say a uniform or normal distribution) be able to use the trained KDE to 'predict' how many of the data points from the given data distribution belong to the target bimodal distribution.
I am using Python 3.8 and sklearn 0.22.
Thanks!