I have large 2D/3D NumPy arrays of binary values. 1s represent boundaries and 0s represent regions. Accompanied by a step size array that indicates the size of step in each dimension.
I am looking for an efficient program that can find (one of) the nearest boundary element to a given element. Distance is euclidean distance.
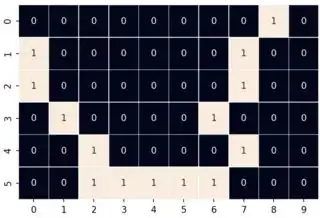
A 2D example:
import seaborn as sns
import numpy as np
step_size = np.array([5,5]) # size of step in each dimension
arr = np.array([[0,0,0,0,0,0,0,0,1,0],[1,0,0,0,0,0,0,1,0,0],[1,0,0,0,0,0,0,1,0,0],[0,1,0,0,0,0,1,0,0,0],[0,0,1,0,0,0,0,1,0,0],[0,0,1,1,1,1,1,0,0,0]])
sns.heatmap(arr, annot=True, cbar=False, linewidths=.3)
a = (0,2) # a given element index
b = (1,0) # nearest boundary element index, which is to be found by the program
a_coor = np.multiply(np.array(a), step_size) # array([0,10])
b_coor = np.multiply(np.array(b), step_size) # array([5,0])
distance = np.linalg.norm(a_coor-b_coor) # 11.18