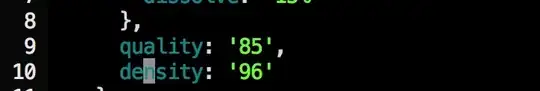
So i was trying this and I find it really unfeasible. I am not that aware about smart ways to do the following. Can somebody help ? Also inputs of lists are quite big.
This task was to create an image from the values generated by me. center_star contains list of [x,y] pairs which are centers of various point like objects.
1800 value represents that image to be generated is of 1800x1800 pixel.
Sigma variable has value 2 by default.
final=[[0]*1800]*1800
for i in range(len(center_stars)):
xi=center_stars[i][0]
yi=center_stars[i][1]
print(i)
for j in range(1800):
for k in range(1800):
final[j][k]+=gauss_amplitude[i]*(math.e**((-1*((xi-j)**2+(yi-k)**2))/2*sigma*sigma))
Is there a smarter way to save time using some of the numpy operation and execute this piece of code in less time?