I want to plot the correlation between several factors in my data set. If possible, I'd like to try to add error bars or whiskers to these plotted values.
Prior to calculating the values, I first want to group them according to the values in one of the columns. I would like to use a tidyverse solution if possible. I am able to achieve half of this using cor(), but I don't know how to add an additional column that contains the p value.
I think the iris data set demonstrates what I'd like to do fairly well. The actual data uses a time series along the x axis. I have spearman specified because that's the correlation used in my analysis, not because it's the correct choice in the iris data set. I've seen some other posts suggesting the use of cor.test and pulling the values from that, but I'm not sure how that would be applied back to the bar chart to use as error bars. Here's the code to create the basic bar chart below.
Edit I have changed my example from using the mtcars data set to the iris data set as I think it better reflects my data. While the initial answer to the question by jay.sf worked with the mtcars set and was greatly appreciated, it didn't work with my data set and the iris set threw the same errors I was having. Additionally, I hadn't stated this in the original, but a tidyverse solution is preferable, but not necessary.
I think that the answer I'm looking for might be contained here, but I'm still trying to work out the details: https://dominicroye.github.io/en/2019/tidy-correlation-tests-in-r/.
iristest <- iris %>%
group_by(Species) %>%
summarise(COR = cor(Sepal.Length,Sepal.Width, method = "spearman", use="complete.obs"))
ggplot(data = iristest) +
aes(x = Species, y = COR) +
geom_bar(stat = "identity") +
theme_minimal()
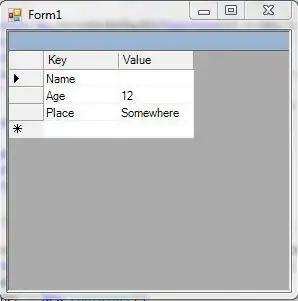
As it is, iristest provides this output:
Species COR
1 setosa 0.7553375
2 versicolor 0.5176060
3 virginica 0.4265165
I think ideally I'd like the output to have the p values added after the COR column.
Species COR p-value
1 setosa 0.7553375 ###
2 versicolor 0.5176060 ###
3 virginica 0.4265165 ###