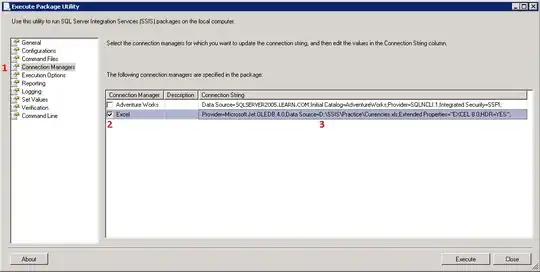
I created a heatmap based on spearman's correlation matrix using seaborn clustermap as folowing: I want to paint the dendrogram. I want the dendrogram to look like this: dendrogram but on the heatmap
I created a dict of colors as folowing and got an error:
def assign_tree_colour(name,val_dict,coding_names_df):
ret = None
if val_dict.get(name, '') == 'Group 1':
ret = "(0,0.9,0.4)" #green
elif val_dict.get(name, '') == 'Group 2':
ret = "(0.6,0.1,0)" #red
elif val_dict.get(name, '') == 'Group 3':
ret = "(0.3,0.8,1)" #light blue
elif val_dict.get(name, '') == 'Group 4':
ret = "(0.4,0.1,1)" #purple
elif val_dict.get(name, '') == 'Group 5':
ret = "(1,0.9,0.1)" #yellow
elif val_dict.get(name, '') == 'Group 6':
ret = "(0,0,0)" #black
else:
ret = "(0,0,0)" #black
return ret
def fix_string(str):
return str.replace('"', '')
external_data3 = [list(z) for z in coding_names_df.values]
external_data3 = {fix_string(z[0]): z[3] for z in external_data3}
tree_label = list(df.index)
tree_label = [fix_string(x) for x in tree_label]
tree_labels = { j : tree_label[j] for j in range(0, len(tree_label) ) }
tree_colour = [assign_tree_colour(label, external_data3, coding_names_df) for label in tree_labels]
tree_colors = { i : tree_colour[i] for i in range(0, len(tree_colour) ) }
sns.set(color_codes=True)
sns.set(font_scale=1)
g = sns.clustermap(df, cmap="bwr",
vmin=-1, vmax=1,
yticklabels=1, xticklabels=1,
cbar_kws={"ticks":[-1,-0.5,0,0.5,1]},
figsize=(13,13),
row_colors=row_colors,
col_colors=col_colors,
method='average',
metric='correlation',
tree_kws=dict(colors=tree_colors))
g.ax_heatmap.set_xlabel('Genus')
g.ax_heatmap.set_ylabel('Genus')
for label in Group.unique():
g.ax_col_dendrogram.bar(0, 0, color=lut[label],
label=label, linewidth=0)
g.ax_col_dendrogram.legend(loc=9, ncol=7, bbox_to_anchor=(0.26, 0., 0.5, 1.5))
ax=g.ax_heatmap
File "<ipython-input-64-4bc6be89afe3>", line 11, in <module>
tree_kws=dict(colors=tree_colors))
File "C:\Users\rotemb\AppData\Local\Continuum\anaconda3\lib\site-packages\seaborn\matrix.py", line 1391, in clustermap
tree_kws=tree_kws, **kwargs)
File "C:\Users\rotemb\AppData\Local\Continuum\anaconda3\lib\site-packages\seaborn\matrix.py", line 1208, in plot
tree_kws=tree_kws)
File "C:\Users\rotemb\AppData\Local\Continuum\anaconda3\lib\site-packages\seaborn\matrix.py", line 1054, in plot_dendrograms
tree_kws=tree_kws
File "C:\Users\rotemb\AppData\Local\Continuum\anaconda3\lib\site-packages\seaborn\matrix.py", line 776, in dendrogram
return plotter.plot(ax=ax, tree_kws=tree_kws)
File "C:\Users\rotemb\AppData\Local\Continuum\anaconda3\lib\site-packages\seaborn\matrix.py", line 692, in plot
**tree_kws)
File "C:\Users\rotemb\AppData\Local\Continuum\anaconda3\lib\site-packages\matplotlib\collections.py", line 1316, in __init__
colors = mcolors.to_rgba_array(colors)
File "C:\Users\rotemb\AppData\Local\Continuum\anaconda3\lib\site-packages\matplotlib\colors.py", line 294, in to_rgba_array
result[i] = to_rgba(cc, alpha)
File "C:\Users\rotemb\AppData\Local\Continuum\anaconda3\lib\site-packages\matplotlib\colors.py", line 177, in to_rgba
rgba = _to_rgba_no_colorcycle(c, alpha)
File "C:\Users\rotemb\AppData\Local\Continuum\anaconda3\lib\site-packages\matplotlib\colors.py", line 240, in _to_rgba_no_colorcycle
raise ValueError("Invalid RGBA argument: {!r}".format(orig_c))
ValueError: Invalid RGBA argument: 0
Any help on this would be greatly appreciated! Tnx!