I'd like to have different annotations (e.g., p-values) on each facet (one for each slope for my actual plot—6 total). I think I've read all the posts on annotating facets, the most useful being of course the main Annotating text on individual facet in ggplot2. But in my situation, it's throwing errors.
I'm using the interactions package, which provides an editable ggplot object, but brings up other issues. Here a minimally reproducible example using mtcars.
# Create the model
mod1 <- lm(wt ~ am * drat * vs, data = mtcars)
# Make the plot
require(interactions)
(p <- interact_plot(mod1,pred="am",modx="drat",mod2="vs"))
# Make annotations dataframe
(dat_text <- data.frame(
text = c("p-value 1", "p-value 2"),
vs = c(0, 1)))
# Add annotations to dataframe
require(ggplot2)
p + geom_text(
data = dat_text,
mapping = aes(x = -Inf, y = -Inf, label = text),
hjust = -0.1,
vjust = -1
)
This gives: Error in FUN(X[[i]], ...) : object 'modx_group' not found. Also same error with 'drat' not found. I'm not too sure how to go about this error (e.g., what values to set them to), so I've tried adding these columns to the dataframe like so:
# Make annotations dataframe
(dat_text <- data.frame(
text = c("p-value 1", "p-value 2"),
vs = c(0, 1),
modx_group = c("-1 SD", "+ 1 SD"), # Here ***
drat = c(-1,1))) # Here ***
# Add annotations to dataframe
p + geom_text(
data = dat_text,
mapping = aes(x = -Inf, y = -Inf, label = text),
hjust = -0.1,
vjust = -1
)
But this gives: Insufficient values in manual scale. 4 needed but only 3 provided. Setting modx_group and drat to NA or NA_real_ or even 0, as indicated in this other post, brings up yet another error: Discrete value supplied to continuous scale.
I can't understand these errors in the current context. I suspect, of course, it has to do with the interactions plot object being funky. There's also probably something obvious I'm doing wrong but can't see. Any help would be appreciated!
Edit
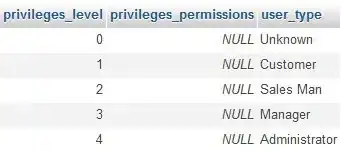
Based on @stefan's answer, I was able to create the desired output for my more complicated design (with 6 p-values, one for each slope, and specific positions for each annotation), as below.
<!-- language-all: lang-r -->
# Create the model
mod1 <- lm(wt ~ am * drat * vs, data = mtcars)
# Make the plot
require(interactions)
#> Loading required package: interactions
(p <- interact_plot(mod1,pred="am",modx="drat",mod2="vs"))
# Make annotations dataframes
dat_text <- data.frame(
text = c("p-value 3", "p-value 6", "p-value 2", "p-value 5", "p-value 1", "p-value 4"),
mod2_group = c("vs = 0", "vs = 1", "vs = 0", "vs = 1", "vs = 0", "vs = 1"),
x = c(0.5, 0.5, 0.5, 0.5, 0.5, 0.5),
y = c(3, 2.5, 3.5, 2.75, 4, 3))
# Add annotations to dataframe
require(ggplot2)
#> Loading required package: ggplot2
p + geom_text(data = dat_text,
mapping = aes(x = x, y = y, label = text),
inherit.aes = FALSE)
Created on 2020-06-10 by the reprex package (v0.3.0)