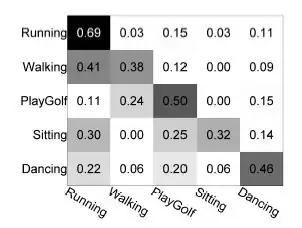
How to get the coordinates of the big rectangles that line on the diagonal.
For example yellow [0,615], [615,1438], [1438,1526]
import numpy as np;
import pandas as pd
from sklearn.metrics.pairwise import cosine_similarity
df = pd.DataFrame(array) # array is image numpy
df.shape #(1526, 360)
s = cosine_similarity(df) #(1526, 1526)
plt.matshow(s)
i try get peaks in first row, but have noise information
speak = 1-s[0]
peaks, _ = find_peaks(speak, distance=160, height=0.1)
print(peaks, len(peaks))
np.diff(peaks)
plt.plot(speak)
plt.plot(peaks, speak[peaks], "x")
plt.show()
Update, add another example And upload to colab full script https://colab.research.google.com/drive/1hyDIDs-QjLjD2mVIX4nNOXOcvCZY4O2c?usp=sharing