We can feed the values to HeatMap in folium as weights (see the data parameter in documentation).
Here's an example. I've divided values in the bottom half of the box by 5 to demonstrate that despite the relatively uniform density of data points, the heat map clearly shows higher magnitudes in the upper half of the picture:
import pandas as pd
import numpy as np
import folium
from folium.plugins import HeatMap
import matplotlib as mpl
# parameters
n = 250 # number of points
lat0 = 40.7 # coordinates will be generated uniformly with
lon0 = -73.9 # lat0 - eps <= lat < lat0 + eps
eps = 0.1 # lon0 - eps <= lon < lon0 + eps
v_min, v_max = 0, 100 # min, max values
# generating values
lat = np.random.uniform(lat0 - eps, lat0 + eps, n)
lon = np.random.uniform(lon0 - eps, lon0 + eps, n)
value = numpy.random.uniform(v_min, v_max, n)
df = pandas.DataFrame({'lat': lat, 'lon': lon, 'value': value})
# to demonstrate the effect of weights on the heatmap,
# we'll divide values below the center of the box by K = 5
K = 5
df.loc[df['lat'] < lat0, 'value'] /= K
# plotting the map, both the points themselves and the heatmap
m = folium.Map(location = [lat0, lon0], tiles="OpenStreetMap",
zoom_start=11, width=400, height=400)
for elt in list(zip(df.lat, df.lon, df.value)):
folium.Circle(elt[:2], color="white", radius=elt[2]).add_to(m)
# df.values used here is a (250, 3) numpy.ndarray
# with (lat, lon, weight) for each data point
HeatMap(data=df.values, min_opacity=0.1).add_to(m)
m
Output:
Update:
Here's a slightly different approach, not using the built-in HeatMaps (which is probably currently a better option in light of https://github.com/python-visualization/folium/issues/1271, as mentioned by @agenis).
We first transform our data, making the grid of squares, where value for each square is the average of original data point values lying within that cell (so it does not depend on the number of points within that cell, only their values).
Then we can visualise those squares by plotting GeoJson polygons on the map. Here's an example:
# define the size of the square
step = 0.02
# calculate values for the grid
x = df.copy()
x['lat'] = np.floor(x['lat'] / step) * step
x['lon'] = np.floor(x['lon'] / step) * step
x = x.groupby(['lat', 'lon'])['value'].mean()
x /= x.max()
x = x.reset_index()
# geo_json returns a single square
def geo_json(lat, lon, value, step):
cmap = mpl.cm.RdBu
return {
"type": "FeatureCollection",
"features": [
{
"type": "Feature",
"properties": {
'color': 'white',
'weight': 1,
'fillColor': mpl.colors.to_hex(cmap(value)),
'fillOpacity': 0.5,
},
"geometry": {
"type": "Polygon",
"coordinates": [[
[lon, lat],
[lon, lat + step],
[lon + step, lat + step],
[lon + step, lat],
[lon, lat],
]]}}]}
# generating a map...
m = folium.Map(location=[lat0, lon0], zoom_start=11, width=400, height=400)
# ...with squares...
for _, xi in x.iterrows():
folium.GeoJson(geo_json(xi['lat'], xi['lon'], xi['value'], step),
lambda x: x['properties']).add_to(m)
# ...and the original points
for elt in list(zip(df.lat, df.lon, df.value)):
folium.Circle(elt[:2], color="white", radius=elt[2]).add_to(m)
m
Output:
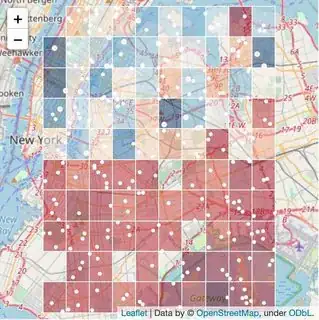
Update (2):
Here's a version with interpolation on the grid using griddata:
import pandas as pd
import numpy as np
import folium
from scipy.interpolate import griddata
# parameters
n = 250 # number of points
lat0 = 40.7
lon0 = -73.9
eps = 0.1
v_min, v_max = 0, 100 # min, max values
# generating values
lat = np.random.normal(lat0, eps, n)
lon = np.random.normal(lon0, eps, n)
value = numpy.random.uniform(v_min, v_max, n)
# set up the grid
step = 0.02
xi, yi = np.meshgrid(
np.arange(lat.min() - step/2, lat.max() + step/2, step),
np.arange(lon.min() - step/2, lon.max() + step/2, step),
)
# interpolate and normalize values
zi = griddata((lat, lon), value, (xi, yi), method='linear')
zi /= np.nanmax(zi)
g = np.stack([
xi.flatten(),
yi.flatten(),
zi.flatten(),
], axis=1)
# geo_json returns a single square
def geo_json(lat, lon, value, step):
cmap = mpl.cm.RdBu
return {
"type": "FeatureCollection",
"features": [
{
"type": "Feature",
"properties": {
'color': 'white',
'weight': 1,
'fillColor': mpl.colors.to_hex(cmap(value)),
'fillOpacity': 0.5,
},
"geometry": {
"type": "Polygon",
"coordinates": [[
[lon - step/2, lat - step/2],
[lon - step/2, lat + step/2],
[lon + step/2, lat + step/2],
[lon + step/2, lat - step/2],
[lon - step/2, lat - step/2],
]]}}]}
# generating a map...
m = folium.Map(location=[lat0, lon0], zoom_start=9, width=400, height=400)
# ...with squares...
for gi in g:
if ~np.isnan(gi[2]):
folium.GeoJson(geo_json(gi[0], gi[1], gi[2], step),
lambda x: x['properties']).add_to(m)
# ...and the original points
for elt in list(zip(lat, lon, value)):
folium.Circle(elt[:2], color='white', radius=elt[2]).add_to(m)
m
Output: