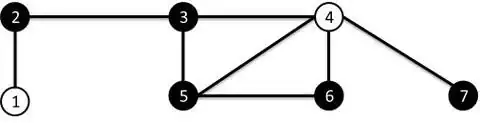
I have a Networkx graph like the following image (image source)
I perform edge attacks and observe the change in values at the node of the resulting subgraph.
Example, If I attack edge (a,2): edge (a, 2) and (2, 1) will be removed. To explain a bit, when edge (a, 2) is attacked the node 2 will have a degree < 2. So the edge that's connected to node 2 is also removed.
The above attack results in a subgraph
Each time an edge is attacked, the value of the terminal node labelled e observed over time changes. Let's say I perform 5 (attack = 5) attacks, I have a time x attack matrix (time=25, attack=5) that stores the time-series data of node e.
I would like to ask for suggestions on how to visualize the effect of these attacks on the value of node e changing over time.
EDIT:
What information do you want to be able to see or identify from your visualizations?
I want to see the attack on which edge has the maximum effect on the time course value observed at e. We could imagine this to be a transportation network and the values at node reflect the amount of a product that has reached the location/node. From the source node b, the goods are transported to target node e. The observation made is the change in node values after an edge is attacked and no observation of the edge value is available.
Please find the code that is used to attack edges
import networkx as nx
import matplotlib.pyplot as plt
def attack(G):
print(G.edges())
for i, edge in enumerate(G.edges()):
no_attack = [(6, 9), (3, 16)]
if edge not in no_attack:
data = {}
print(f'attacking edge {edge}')
H = G.copy()
# attack an edge
H.remove_edges_from(ebunch=[edge])
n = len(G.nodes)
retain_node_ids = [9, 3]
H.add_edges_from([(u, v) for u in retain_node_ids for v in (n+1, n+2)])
# remove nodes with degree < 2
H = nx.k_core(H, k=2)
H.remove_nodes_from([n + 1, n + 2])
# graph_utils_py.draw_graph3d(H, fig=2, show=True)
# H = nx.convert_node_labels_to_integers(H, first_label=1, ordering='default', label_attribute=None)
# delete connected nodes and edges
diff_nodes = set(G.nodes()).difference(H.nodes())
diff_edges = {e for e in G.edges() for n in diff_nodes if n in e}
print(f"deleting connected nodes {diff_nodes} ...")
print(f"deleting connected edges {diff_edges} ...")
data['diff_nodes'] = list(diff_nodes)
data['diff_edges'] = list(diff_edges)
data['edge'] = edge
if __name__ == '__main__':
n = 20
G = nx.gnm_random_graph(n=20, m=30, seed=1)
# nx.draw(G, with_labels=True)
# plt.show()
retain_node_ids = [11, 4]
G.add_edges_from([(u, v) for u in retain_node_ids for v in (n, n + 1)])
G = nx.k_core(G, k=2)
G.remove_nodes_from([n, n + 1])
# nx.draw(G, with_labels=True)
# plt.show()
G = nx.convert_node_labels_to_integers(G, first_label=1, ordering='default', label_attribute=None)
nx.draw(G, with_labels=True)
plt.show()
attack(G)
EDIT2: The answer posted below suggests visualizing the edge attacks by varying the opacity and setting different color schemes. Unfortunately, this doesn't help. One has to create a different image for each attack. I am still looking for other suggestions.
EDIT3: Clarifying a bit more on what exactly I want to visualize to keep things simple.
I'm looking for an interactive graph like the following.
One could click the edge that is attacked and the LHS plot will display the observation made at the target node. The dashed lines are the edges that are affected (stored in variable diff_edges in the code) as a result of an attack on a given edge (stored in variable edge).
If there are overlaps in the edges that are affected after attacking a link, we could display it as multiple lines with the corresponding color mappings. An interactive graph will help the user pick and choose the edge attacks to compare the observation at node e. The edges that are attacked can be displayed by varying the opacity/ line style/ color.
EDIT4: The answer posted below helps. But there is a problem when the impacted edges overlap.
Example, attack(H, (6, 4), color='red') attack(H, (5, 4), color='yellow')
gives
The colors overlap and it's hard to visualize. If we can draw the impacted edges next to each other, without overlapping, as shown in the image posted above in edit3 that will be good.