Description
The motivation for this question is from clinical/epidemiological research, wherein studies often enroll patients and then follow them for variable lengths of time.
The distribution of age at study entry is often of interest and is easily assessed, however there is occasional interest in the distribution of age at any time during the study.
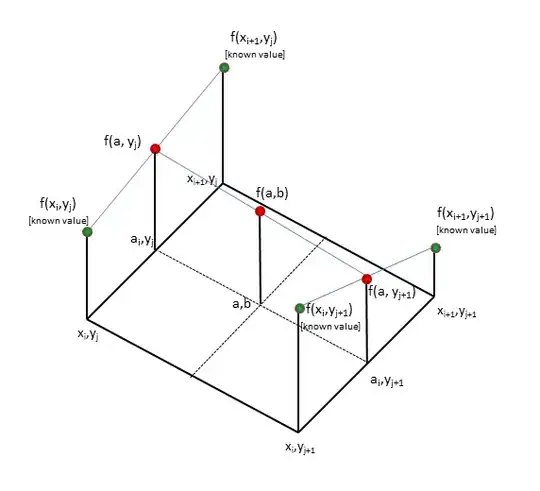
My question is, is there a method for estimating such a density from interval data such as [age_start, age_stop] without expansion of the data as below? The long-format method seems inelegant, to say nothing of its memory usage!
Reproducible example using data from the survival package
#### Prep Data ###
library(survival)
library(ggplot2)
library(dplyr)
data(colon, package = 'survival')
# example using the colon dataset from the survival package
ccdeath <- colon %>%
# use data on time to death (not recurrence)
filter(etype == 2) %>%
# age at end of follow-up (death or censoring)
mutate(age_last = age + (time / 365.25))
#### Distribution Using Single Value ####
# age at study entry
ggplot(ccdeath, aes(x = age)) +
geom_density() +
labs(title = "Fig 1.",
x = "Age at Entry (years)",
y = "Density")
#### Using Person-Month Level Data ####
# create counting-process/person-time dataset
ccdeath_cp <- survSplit(Surv(age, age_last, status) ~ .,
data = ccdeath,
cut = seq(from = floor(min(ccdeath$age)),
to = ceiling(max(ccdeath$age_last)),
by = 1/12))
nrow(ccdeath_cp) # over 50,000 rows
# distribution of age at person-month level
ggplot(ccdeath_cp, aes(x = age)) +
geom_density() +
labs(title = "Figure 2: Density based on approximate person-months",
x = "Age (years)",
y = "Density")
#### Using Person-Day Level Data ####
# create counting-process/person-time dataset
ccdeath_cp <- survSplit(Surv(age, age_last, status) ~ .,
data = ccdeath,
cut = seq(from = floor(min(ccdeath$age)),
to = ceiling(max(ccdeath$age_last)),
by = 1/365.25))
nrow(ccdeath_cp) # over 1.5 million rows!
# distribution of age at person-month level
ggplot(ccdeath_cp, aes(x = age)) +
geom_density() +
labs(title = "Figure 3: Density based on person-days",
x = "Age (years)",
y = "Density")
Note: while I tagged this question with "survival" because I thought it would attract people familiar with this area, I am not interested in time-to-event here, just the overall age distribution of all time under study.