I'm a bit puzzled: I calculated PCAs of the same dataset. Here's the workflow:
- Orange 3.26: Read a .csv, PCA on 4 PCs (normalized variables), scatterplot
- scikit-learn: Read the same .csv, standardizing of numerical values
(StandardScaler(with_mean=True,with_std=True)), PCA (copy=True, iterated_power='auto', n_components=4, random_state=None, svd_solver='auto', tol=0.0, whiten=False)
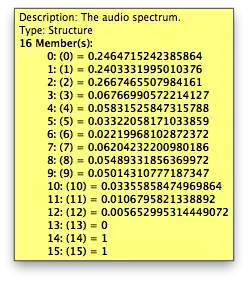
The results differ in the numerical values for the single PCs:
Orange 3.26:
Here is my code for scikit-learn-fu:
I have a pd.DataFrame, shape is (268,16). In a first step I slice the dataframe in two daframes:
- A1: containing all rows and all features; shape(268, 13)
- B1: containing the targets and the ID of every row; shape(268,3)
In a next step I standardize dataframe A1 with StandardScaler from sklearn.preprocessing:
a1 = StandardScaler(with_mean=True,with_std=True).fit_transform(A1)
The next step is the PCA:
pca1 = PCA(n_components=4)
principalComponents1 = pca1.fit_transform(a1)
The outputs are the scores and loadings - nothing special.
Perhaps a difference in normalization of the initial dataset? Any suggestions?