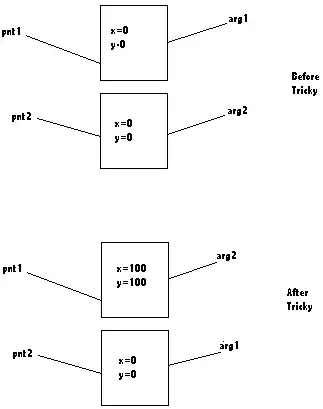
I need to align the x axes on these plots, I think if I could change the width so they had the same width would work but I haven't been able to do it.

my data looks like this:
> x<-as.data.frame(table(casosbog$fis))
> dput(x[1:10,c('Var1','Freq')])
structure(list(Var1 = structure(1:10, .Label = c("2020-02-27",
"2020-02-28", "2020-03-01", "2020-03-04", "2020-03-05", "2020-03-06",
"2020-03-07", "2020-03-08", "2020-03-09", "2020-03-10", "2020-03-11",
"2020-03-12", "2020-03-13", "2020-03-14", "2020-03-15", "2020-03-16",
"2020-03-17", "2020-03-18", "2020-03-19", "2020-03-20", "2020-03-21",
"2020-03-22", "2020-03-23", "2020-03-24", "2020-03-25", "2020-03-26",
"2020-03-27", "2020-03-28", "2020-03-29", "2020-03-30", "2020-03-31",
"2020-04-01", "2020-04-02", "2020-04-03", "2020-04-04", "2020-04-05",
"2020-04-06", "2020-04-07", "2020-04-08", "2020-04-09", "2020-04-10",
"2020-04-11", "2020-04-12", "2020-04-13", "2020-04-14", "2020-04-15",
"2020-04-16", "2020-04-17", "2020-04-18", "2020-04-19", "2020-04-20",
"2020-04-21", "2020-04-22", "2020-04-23", "2020-04-24", "2020-04-25",
"2020-04-26", "2020-04-27", "2020-04-28", "2020-04-29", "2020-04-30",
"2020-05-01", "2020-05-02", "2020-05-03", "2020-05-04", "2020-05-05",
"2020-05-06", "2020-05-07", "2020-05-08", "2020-05-09", "2020-05-10",
"2020-05-11", "2020-05-12", "2020-05-13", "2020-05-14", "2020-05-15",
"2020-05-16", "2020-05-17", "2020-05-18", "2020-05-19", "2020-05-20",
"2020-05-21", "2020-05-22", "2020-05-23", "2020-05-24", "2020-05-25",
"2020-05-26", "2020-05-27", "2020-05-28", "2020-05-29", "2020-05-30",
"2020-05-31", "2020-06-01", "2020-06-02", "2020-06-03", "2020-06-04",
"2020-06-05", "2020-06-06", "2020-06-07", "2020-06-08", "2020-06-09",
"2020-06-10", "2020-06-11", "2020-06-12", "2020-06-13", "2020-06-14",
"2020-06-15", "2020-06-16", "2020-06-17", "2020-06-18", "2020-06-19",
"2020-06-20", "2020-06-21", "2020-06-22", "2020-06-23", "2020-06-24",
"2020-06-25", "2020-06-26", "2020-06-27", "2020-06-28", "2020-06-29",
"2020-06-30", "2020-07-01", "2020-07-02", "2020-07-03", "2020-07-04",
"2020-07-05", "2020-07-06", "2020-07-07", "2020-07-08", "2020-07-09",
"2020-07-10", "2020-07-11", "2020-07-12", "2020-07-13", "2020-07-14",
"2020-07-15", "2020-07-16", "2020-07-17", "2020-07-18", "2020-07-19",
"2020-07-20", "2020-07-21", "2020-07-22", "2020-07-23", "2020-07-24",
"2020-07-25", "2020-07-26", "2020-07-27", "2020-07-28", "2020-07-29",
"2020-07-30", "2020-07-31", "2020-08-01", "2020-08-02", "2020-08-03",
"2020-08-04", "2020-08-05", "2020-08-06", "2020-08-07", "2020-08-08",
"2020-08-09", "2020-08-10", "2020-08-11", "2020-08-12", "2020-08-13",
"2020-08-14", "2020-08-15", "2020-08-16", "2020-08-17", "2020-08-18",
"2020-08-19", "2020-08-20", "2020-08-21", "2020-08-22", "2020-08-23",
"2020-08-24", "2020-08-25", "2020-08-26", "2020-08-27", "2020-08-28",
"2020-08-29", "2020-08-30", "2020-08-31", "2020-09-01", "2020-09-02",
"2020-09-03", "2020-09-04", "2020-09-05", "2020-09-06", "2020-09-07",
"2020-09-08", "2020-09-09", "2020-09-10", "2020-09-11", "2020-09-12",
"2020-09-13", "2020-09-14", "2020-09-15", "2020-09-16", "2020-09-17"
), class = "factor"), Freq = c(1L, 1L, 3L, 1L, 5L, 6L, 10L, 5L,
5L, 11L)), row.names = c(NA, 10L), class = "data.frame")
and
> movilcolbog[1:10,c('date','MovilidadProm')]
date MovilidadProm
1 2020-02-15 7.0000000
2 2020-02-16 4.6666667
3 2020-02-17 5.3333333
4 2020-02-18 7.5000000
5 2020-02-19 2.6666667
6 2020-02-20 -0.3333333
7 2020-02-21 5.0000000
8 2020-02-22 1.8333333
9 2020-02-23 -7.3333333
10 2020-02-24 3.5000000
as you can see both dataframes have different starting dates and dates frequencies, the rest of the code is as follows
library(ggplot2)
library(gridExtra)
plot2<-ggplot(movilcolbaq,aes(date,MovilidadProm))+geom_line(aes(color="Mov. BAQ"),size=1)+
geom_line(data=movilcolbog,aes(color="Mov. BOG"),size=1)+
labs(color="Legend text")+
labs(title='Movilidad promedio (con respecto a enero-febrero) BOG, BAQ',x='',y='Cambio de movilidad en %')+
theme(legend.title=element_blank())
plot1<-ggplot(casosbog,aes(x=fis,fill='Casos Bog'))+geom_bar()+
geom_bar(data=casosbar,aes(x=fis,fill='Casos Bar'),alpha=0.7)+
scale_colour_manual("", values = c("Casos Bog"="red", "Casos Baq"="blue"))+
theme(legend.title=element_blank())+labs(x='Fecha de inicio de síntomas',
y='Casos',
title='Fecha de inicio síntomas BAQ vs BOG vs MED',
subtitle='Fuentes: INS y www.google.com/covid19/mobility/')
grid.arrange(plot1,plot2)
I've tried changing the xlim but it hasn't worked