Current situation: I'm trying to extract segments from an image. Thanks to openCV's findContours() method, I now have a list of 8-connected point for every contours. However, these lists are not directly usable, because they contain a lot of duplicates.
The problem: Given a list of 8-connected points, which can contain duplicates, extract segments from it.
Possible solutions:
- At first, I used openCV's
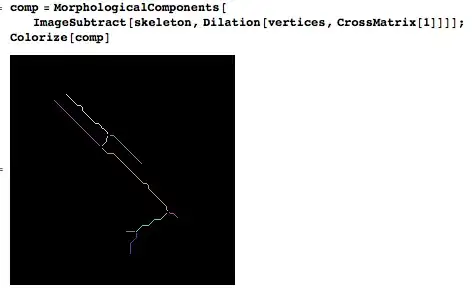
approxPolyDP()method. However, the results are pretty bad... Here is the zoomed contours:
Here is the result of approxPolyDP(): (9 segments! Some overlap)

but what I want is more like:
It's bad because approxPolyDP() can convert something that "looks like several segments" in "several segments". However, what I have is a list of points that tend to iterate several times over themselves.
For example, if my points are:
0 1 2 3 4 5 6 7 8
9
Then, the list of point will be 0 1 2 3 4 5 6 7 8 7 6 5 4 3 2 1 9... And if the number of points become large (>100) then the segments extracted by approxPolyDP() are unfortunately not duplicates (i.e : they overlap each other, but are not strictly equal, so I can't just say "remove duplicates", as opposed to pixels for example)
- Perhaps, I've got a solution, but it's pretty long (though interesting). First of all, for all 8-connected list, I create a sparse matrix (for efficiency) and set the matrix values to 1 if the pixel belongs to the list. Then, I create a graph, with nodes corresponding to pixels, and edges between neighbouring pixels. This also means that I add all the missing edges between pixels (complexity small, possible because of the sparse matrix). Then I remove all possible "squares" (4 neighbouring nodes), and this is possible because I am already working on pretty thin contours. Then I can launch a minimal spanning tree algorithm. And finally, I can approximate every branch of the tree with openCV's
approxPolyDP()
To sum up: I've got a tedious method, that I've not yet implemented as it seems error-prone. However, I ask you, people at Stack Overflow: are there other existing methods, possibly with good implementations?
Edit: To clarify, once I have a tree, I can extract "branches" (branches start at leaves or nodes linked to 3 or more other nodes) Then, the algorithm in openCV's approxPolyDP() is the Ramer–Douglas–Peucker algorithm, and here is the Wikipedia picture of what it does:

With this picture, it is easy to understand why it fails when points may be duplicates of each other
Another edit: In my method, there is something that may be interesting to note. When you consider points located in a grid (like pixels), then generally, the minimal spanning tree algorithm is not useful because there are many possible minimal trees
X-X-X-X
|
X-X-X-X
is fundamentally very different from
X-X-X-X
| | | |
X X X X
but both are minimal spanning trees
However, in my case, my nodes rarely form clusters because they are supposed to be contours, and there is already a thinning algorithm that runs beforehand in the findContours().
Answer to Tomalak's comment:

If DP algorithm returns 4 segments (the segment from the point 2 to the center being there twice) I would be happy! Of course, with good parameters, I can get to a state where "by chance" I have identical segments, and I can remove duplicates. However, clearly, the algorithm is not designed for it.
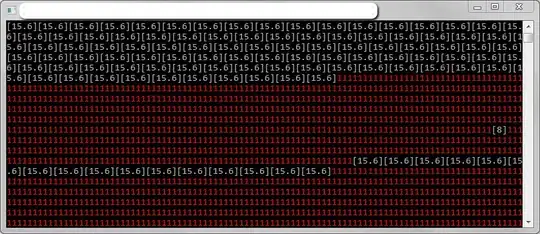
Here is a real example with far too many segments: