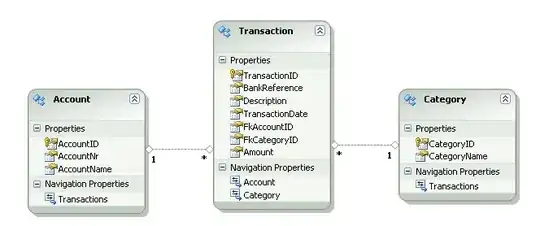
I'm trying to load data from an excel sheet and then plot all on the same plot but I am a little inexperienced with plotting multiple lines on a single plot. Each column is Time Elapsed and the corresponding Residual Act. and I have multiple columns named the same. Time is different in each column hence having multiple time columns. Right now the code just outputs 4 separate plots. Can someone tell me how to do this without overly complicating myself, I have to plot multiple files in the future and would like an easy way.
import pandas as pd
import matplotlib.pyplot as plt
HD = pd.read_excel('C:\\Users\\azizl\\Desktop\\HDPD_Data.xlsx')
HD.plot(x='Time Elapsed', y= 'Residual Act.' , label='24')
HD.plot(x='Time Elapsed.1', y= 'Residual Act..1', label='48')
HD.plot(x='Time Elapsed.2', y= 'Residual Act..2', label='normal')
HD.plot(x='Time Elapsed.3', y= 'Residual Act..3', label='physical')
plt.show()
HD.head()