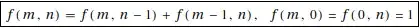I want to plot the predicted probabilities for a multinomial model in R, fitted with the nnet::multinom() function. I have numerical predictors on the log scale.
Even though {ggeffects} should be compatible with multinom(), the plot does not display confidence intervals the same way it does for linear models.
I am new to using to R and to this community, so my apologies if this question is very basic or is missing something essential. Here is a small example:
library(tidyverse)
library(nnet)
library(effects)
library(ggeffects)
df <- data.frame(response = c("1 Better", "1 Better", "1 Better", "2 Medium", "2 Medium", "2 Medium", "3 Worse", "3 Worse", "3 Worse"),
count = c(1000, 2000, 4000, 6000, 10000, 3000, 6000, 5000, 11000))
mod1 <- multinom(response ~ log(count), data = df)
summary(mod1)
effects::effect(mod1, term="log(count)", se=TRUE, confidence.level=.95) %>% plot() # Produces CIs.
ggeffects::ggpredict(mod1, terms = "count") %>% plot() + theme_bw() # No confidence intervals.
If others are looking for alternatives to {ggeffects}, I tried these approaches while looking for a solution:
Using effects::effect(): works, confidence intervals are included but the appearance isn't so customisable.
Combining {ggeffects} and {effects}: See this post on R Studio Community in which confidence intervals from the effects package were combined with ggeffects to create a plot. I got the error
Error in FUN(X[[i]], ...) : object 'L' not found
But it worked for that person.
Using {MNLpred} package and its mnl_pred_ova() : didn't work for me, I think because my predictors are in log scale. I got the following error:
Error in eval(parse(text = paste0("data$", xvari))) : attempt to apply non-function
Using mnlAveEffPlot() function from {DAMisc}: Worked but the plots aren't as customisable as I would like.