So I'm trying to do a very simple boxplot of one continuous variable against one discrete yes/no variable, and for reasons I totally don't understand, I can't get the range bars to display for the "NO" values.
Here's a simplified dataset ... save in your working directory as "femplot.csv"
SEQN,LBXVBZ,smoke
73614,0.206,YES
73616,0.017,NO
73739,0.017,NO
73751,0.135,YES
73763,0.237,YES
73766,0.017,NO
73805,0.19,YES
73848,0.017,NO
73914,0.198,YES
73924,0.017,NO
73938,0.161,YES
73975,0.167,YES
74006,0.031,YES
74007,0.017,NO
74008,0.017,NO
74022,0.147,YES
74046,0.017,NO
74054,0.017,NO
74091,0.156,YES
74101,0.179,YES
74141,0.106,NO
74150,0.115,YES
74154,0.017,NO
74160,0.017,NO
74173,0.035,NO
74180,0.017,NO
74195,0.017,NO
74211,0.017,NO
74221,0.078,YES
Now here's my code ... I'm trying this both using the R-native boxplot function and ggplot, with the same result:
library(ggplot2)
femplot <- read.csv("femplot.csv")
boxplot(LBXVBZ~smoke, data = femplot)
ggplot(data = femplot, aes(x=smoke, y=LBXVBZ))+
stat_boxplot(geom="errorbar", width=0.5, coef = 10)+
geom_boxplot()+
stat_summary(fun = "mean", shape=23, color="red")+
labs(x="Smoker", y="Benzene"),
title = "Distribution of blood benzene levels among smokers/nonsmokers")
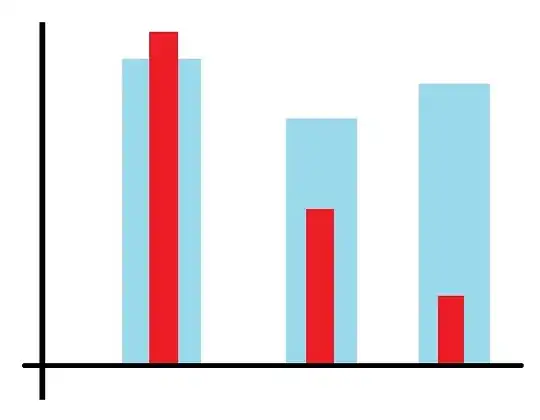
Here's the output. Note that there's a box and range lines on the "YES" values, but none for the "NO". In point of fact the box is vanishingly small for "NO", so that's just fine, but I should still get rangelines, since there are dots to show a range. I haven't bothered to include the image for the standard boxplot output but it's equivalent.