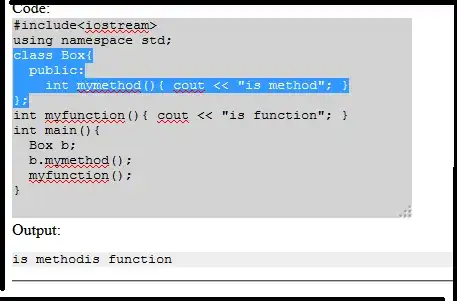
I have an API pull request that returns a .gz file, which R recognizes as a "raw" file. I have not been able to find any R package that can decompress it in-memory from the saved file. I've tried fread(), rawToChar() and unzip().
The structure of the file I need to unzip is below. Specifically req$content.