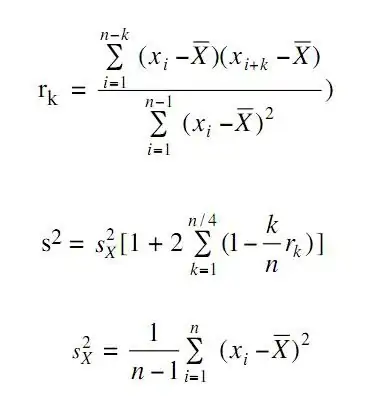Hello stackoverflow Community,
I want to create a line chart for all countries in the data set (x = year, y = BMI). I just want to use R base for visualization. The problem is that R generates the visualization seperatly for each country. I want one visulization for all countries with seperate margins for each country within the visualization.
Thank you for helping.
Dataset: https://github.com/tanaytuncer/LifeExpectancy_BMI Code:
path2 <- "/Users/tanaytuncer/Desktop/Quantitative Datenanalyse/BMI.csv"
data <- read.csv(path2, check.names = FALSE)
data <- data[-1:-3, ]
names(data)[1] <- "country"
data <- data %>%
mutate(across(-country, parse_number)) %>%
gather("year", "BMI", 2:17)
df_BMI4 <- data %>%
select(country, BMI, year)
View(df_BMI4)
par(mfrow=c(50,4), mar(4, 3, 3, 1))
for (i in df_BMI4$country) {
country <- subset(df_BMI4, country == i)
plot(country$year, country$BMI, type="l", main = i, add = TRUE)
}