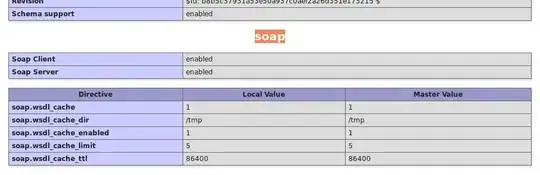
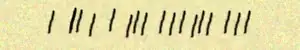I am python beginner and I have one problem I can't solve. I need to make a 2d hystogram using matrix I got in txt, also I can get it in xls. Example of matrix:
4.52 4.54 4.52 4.44 4.34 4.28
5.10 4.92 4.82 4.80 4.66 4.44
6.12 5.80 5.57 5.50 5.15 4.70
6.47 6.54 6.27 6.13 6.21 5.97
8.11 8.73 8.70 8.63 8.84 8.55
I can get it in code:
a = np.loadtxt('matrix.txt')
And then I will have it. So I have a part of code, which make a hystogram like an example, but I don't understand how integrate my matrix in here:
import matplotlib.pyplot as plt
n = 100000
x = np.random.standard_normal(n)
y = 2.0 + 3.0 * x + 4.0 * np.random.standard_normal(n)
xedges = np.linspace(-4, 4, 42)
yedges = np.linspace(-25, 25, 42)
hist, xedges, yedges = np.histogram2d(x, y, (xedges, yedges))
xidx = np.clip(np.digitize(x, xedges), 0, hist.shape[0]-1)
yidx = np.clip(np.digitize(y, yedges), 0, hist.shape[1]-1)
c = hist[xidx, yidx]
plt.scatter(x, y, c=c)
plt.show()
With the help of this code I want to make a hystogram using my matrix, but I don't understand how. I will be thankful.