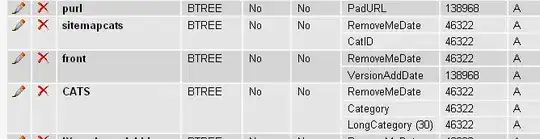
Let the sample df (df1) be,
We can achieve df2 or final data-frame by manipulating the data of df1 in the following manner,
Step 1: Remove all positive numbers including zeros
After Step 1 the sample data should look like,
Step 2: If column4 A row is a negative number and column4 B is blank, then remove the -ve number of column4 A row
Step 3: If column4 A row is blank and column4 B is a negative number, then keep the -ve number of column4 B row
After Steps 1,2 and 3 are done,
Step 4: If both A and B of column4 are negative then,
For each A and B row of column4, check the left-side (LHS) value (for a given month) of the same A and B row of column4
Step 4.1: If either of the LHS values of A or B is a -ve number, then delete the current row value of B column4 and keep the current row value of A column4
After Step 4.1, the sample data should look like this,
Step 4.2:
If the LHS value of A and B column4 is blank, then keep the current row value of B column4 and delete the current row value of A column4
Sample data after Step 4.2 should look like,
Since we see two negative numbers still, we perform Step 4.1 again and then the final data-frame or df2 will look like,
How may I achieve the above using pandas? I was able to achieve till Step 1 but have no idea as to how to proceed further. Any help would be greatly appreciated.
This is the approach that I took,
import pandas as pd
df = pd.read_excel('df1.xlsx', engine='openpyxl')
df.to_pickle('./df1.pkl')
unpickled_df = pd.read_pickle('./df1.pkl')
rem_cols = ['column2', 'column3', 'column5', 'column6', 'column7']
unpickled_df['g'] = unpickled_df.groupby(['column1', 'column4'] ).cumcount()
df1 = unpickled_df.drop(rem_cols, axis=1)
df1 = df1.set_index(['column1','g', 'column4'])
df1.columns = pd.to_datetime(df1.columns, format='%b-%y').strftime('%b-%y')
first_date = df1.columns[0]
df1 = df1.unstack(-1)
df1 = df1.mask(df1.ge(0))
m1 = (df1.xs('A', level=1, axis=1, drop_level=False).notna() &
df1.xs('B', level=1, axis=1, drop_level=False).rename(columns={'B':'A'}, level=1).isna())
m2 = (df1.xs('B', level=1, axis=1, drop_level=False).notna() &
df1.xs('A', level=1, axis=1, drop_level=False).rename(columns={'A':'B'}, level=1).isna())
m = m1.join(m2)
df1 = df1.mask(m)
df2 = df1.groupby(level=1, axis=1).shift(1, axis=1)
mask1 = df1.notna() & df2.isna() & (df1.columns.get_level_values(1) == 'A')[ None, :]
mask1[first_date] = False
mask2 = df1.notna() & df2.notna() & (df1.columns.get_level_values(1) == 'B')[ None, :]
df1 = df1.mask(mask1).mask(mask2).stack(dropna=False)
unpickled_df = unpickled_df[rem_cols + ['column1','g', 'column4']].join(df1, on=['column1','g', 'column4'])
#print(unpickled_df)
Small Test data: df1,
{'column1': ['ABC', 'ABC', 'CDF', 'CDF'], 'column4': ['A', 'B', 'A', 'B'], 'Feb-21': [0, 10, 0, 0], 'Mar-21': [0, 0, 70, 70], 'Apr-21': [-10, -10, -8, 60], 'May-21': [-30, -60, -10, 40], 'Jun-21': [-20, 9, -40, -20], 'Jul-21': [30, -10, 0, -20], 'Aug-21': [-30, -20, 0, -20], 'Sep-21': [0, -15, 0, -20], 'Oct-21': [0, -15, 0, -20]}
df2 (expected output),
{'column1': ['ABC', 'ABC', 'CDF', 'CDF'], 'column4': ['A', 'B', 'A', 'B'], 'Feb-21': [nan, nan, nan, nan], 'Mar-21': [nan, nan, nan, nan], 'Apr-21': [nan, -10.0, nan, nan], 'May-21': [-30.0, nan, nan, nan], 'Jun-21': [nan, nan, nan, -20.0], 'Jul-21': [nan, -10.0, nan, -20.0], 'Aug-21': [-30.0, nan, nan, -20.0], 'Sep-21': [nan, -15.0, nan, -20.0], 'Oct-21': [nan, -15.0, nan, -20.0]}
Test data:
df1
{'column1': ['CT', 'CT', 'NBB', 'NBB', 'CT', 'CT', 'NBB', 'NBB', 'HHH', 'HHH', 'TP1', 'TP1', 'TPR', 'TPR', 'PP1', 'PP1', 'PP1', 'PP1'], 'column2': ['POUPOU', 'POUPOU', 'PRPRP', 'PRPRP', 'STDD', 'STDD', 'STDD', 'STDD', 'STEVT', 'STEVT', 'SYSYS', 'SYSYS', 'SYSYS', 'SYSYS', 'SHW', 'SHW', 'JV', 'JV'], 'column3': ['V', 'CV', 'V', 'CV', 'V', 'CV', 'V', 'CV', 'V', 'CV', 'V', 'CV', 'V', 'CV', 'V', 'CV', 'V', 'CV'], 'column4': ['A', 'B', 'A', 'B', 'A', 'B', 'A', 'B', 'A', 'B', 'A', 'B', 'A', 'B', 'A', 'B', 'A', 'B'], 'column5': [nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan], 'column6': ['BBB', 'BBB', 'CCC', 'CCC', 'BBB', 'BBB', 'BBB', 'BBB', 'VVV', 'VVV', 'CHCH', 'CHCH', 'CHCH', 'CHCH', 'CCC', 'CCC', 'CHCH', 'CHCH'], 'column7': ['Apr-21', 'Apr-21', 'Apr-21', 'Apr-21', 'Apr-21', 'Apr-21', 'Apr-21', 'Apr-21', 'Mar-21', 'Mar-21', 'Mar-21', 'Mar-21', 'Mar-21', 'Mar-21', 'Apr-21', 'Apr-21', 'Mar-21', 'Mar-21'], 'Feb-21': [11655, 0, 0, 0, 121117, 0, 14948, 0, 0, 0, 0, 0, 0, 0, 1838, 0, 0, 0], 'Mar-21': [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, -16474.0, -16474.0, 7000.0, 7000.0, -19946.0, -19946.0, 16084.44444444444, 0.0, 0.0, 0.0], 'Apr-21': [104815.0, 104815.0, 17949.0, 17949.0, 96132.0, 96132.0, 0.0, 0.0, -17001.0, -33475.0, -878.0, 6122.0, 8398.0, -11548.0, -5297.073170731703, -5297.073170731703, -282.0, -282.0], 'May-21': [78260.0, 183075.0, 42557.0, 60506.0, -15265.0, 80867.0, -18.0, -18.0, 21084.0, -12391.0, -1831.0, 4291.0, 2862.0, -8686.0, 5261.25, -35.8231707317027, -369.0, -651.0], 'Jun-21': [-52480.0, 130595.0, -13258.0, 47248.0, -35577.0, 45290.0, 2434.0, 2416.0, 31147.0, 18756.0, -4310.0, -19.0, -4750.0, -13436.0, -92.0, -127.8231707317027, -280.0, -931.0], 'Jul-21': [-174544.0, -43949.0, -38127.0, 9121.0, -124986.0, -79696.0, -9707.0, -7291.0, 13577.0, 32333.0, 0.0, -19.0, -15746.0, -29182.0, 93.0, -34.8231707317027, -319.0, -1250.0], 'Aug-21': [35498.0, -8451.0, -37094.0, -27973.0, 79021.0, -675.0, -1423.0, -8714.0, 32168.0, 64501.0, 0.0, -19.0, 18702.0, -10480.0, 4347.634146341465, 4312.810975609762, -341.0, -1591.0], 'Sep-21': [44195.0, 35744.0, 2039.0, -25934.0, 70959.0, 70284.0, 2816.0, -5898.0, 38359.0, 102860.0, 0.0, -19.0, 18119.0, 7639.0, 5302.222222222219, 9615.033197831981, 0.0, -1591.0], 'Oct-21': [-13163.0, 22581.0, -4773.0, -30707.0, 205080.0, 275364.0, -709.0, -6607.0, -1397.0, 101463.0, 0.0, -19.0, 0.0, 7639.0, -34.0, 9581.033197831981, 0.0, -1591.0]}
df2 (expected output) ,
{'column1': ['CT', 'CT', 'NBB', 'NBB', 'CT', 'CT', 'NBB', 'NBB', 'HHH', 'HHH', 'TP1', 'TP1', 'TPR', 'TPR', 'PP1', 'PP1', 'PP1', 'PP1'], 'column2': ['POUPOU', 'POUPOU', 'PRPRP', 'PRPRP', 'STDD', 'STDD', 'STDD', 'STDD', 'STEVT', 'STEVT', 'SYSYS', 'SYSYS', 'SYSYS', 'SYSYS', 'SHW', 'SHW', 'JV', 'JV'], 'column3': ['V', 'CV', 'V', 'CV', 'V', 'CV', 'V', 'CV', 'V', 'CV', 'V', 'CV', 'V', 'CV', 'V', 'CV', 'V', 'CV'], 'column4': ['A', 'B', 'A', 'B', 'A', 'B', 'A', 'B', 'A', 'B', 'A', 'B', 'A', 'B', 'A', 'B', 'A', 'B'], 'column5': [nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan], 'column6': ['BBB', 'BBB', 'CCC', 'CCC', 'BBB', 'BBB', 'BBB', 'BBB', 'VVV', 'VVV', 'CHCH', 'CHCH', 'CHCH', 'CHCH', 'CCC', 'CCC', 'CHCH', 'CHCH'], 'column7': ['Apr-21', 'Apr-21', 'Apr-21', 'Apr-21', 'Apr-21', 'Apr-21', 'Apr-21', 'Apr-21', 'Mar-21', 'Mar-21', 'Mar-21', 'Mar-21', 'Mar-21', 'Mar-21', 'Apr-21', 'Apr-21', 'Mar-21', 'Mar-21'], 'Feb-21': [nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan], 'Mar-21': [nan, nan, nan, nan, nan, nan, nan, nan, nan, -16474.0, nan, nan, nan, -19946.0, nan, nan, nan, nan], 'Apr-21': [nan, nan, nan, nan, nan, nan, nan, nan, -17001.0, nan, nan, nan, nan, -11548.0, nan, -5297.073170731703, nan, -282.0], 'May-21': [nan, nan, nan, nan, nan, nan, nan, -18.0, nan, -12391.0, nan, nan, nan, -8686.0, nan, -35.8231707317027, -369.0, nan], 'Jun-21': [nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, nan, -19.0, -4750.0, nan, -92.0, nan, -280.0, nan], 'Jul-21': [nan, -43949.0, nan, nan, nan, -79696.0, nan, -7291.0, nan, nan, nan, -19.0, -15746.0, nan, nan, -34.8231707317027, -319.0, nan], 'Aug-21': [nan, -8451.0, nan, -27973.0, nan, -675.0, -1423.0, nan, nan, nan, nan, -19.0, nan, -10480.0, nan, nan, -341.0, nan], 'Sep-21': [nan, nan, nan, -25934.0, nan, nan, nan, -5898.0, nan, nan, nan, -19.0, nan, nan, nan, nan, nan, -1591.0], 'Oct-21': [nan, nan, -4773.0, nan, nan, nan, -709.0, nan, nan, nan, nan, -19.0, nan, nan, nan, nan, nan, -1591.0]}
Note: I have implemented my code on the basis of the Test data provided. The sample data is merely to focus on the columns that are supposed to be manipulated.