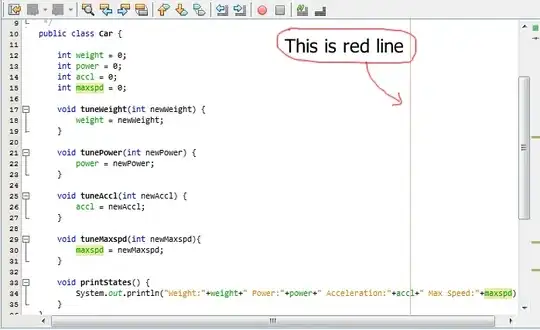
When I run the same R code in my local computer RStudio (R 4.0.2) and on Code Ocean R 4.0.3, I have two different UMAP visualization results and they are mirrored
[ ]
]
I use Seurat 3.2.0 version in both environments and particularly for umap visualization, here is the line:
DimPlot(MU197PDXThgFiltered, reduction = "umap", label = TRUE, label.size = 6, pt.size = 0.5)
I can't give a reproducible example, but maybe someone faced this issue before?