I have this dataset from Kaggle that is JSON.
beer <- fromJSON('recipes_full.txt')
It imports as a giant list. For example, each beer#, I'll start with 0, has a list of 19.
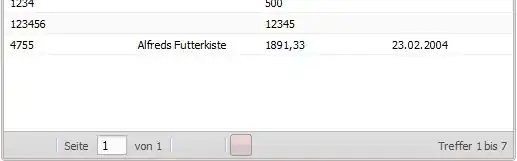 Each of those lists has a single value, except for 4 of them, which are different sized tables, like 8x2, 5x5, 2x4, ect.
Each of those lists has a single value, except for 4 of them, which are different sized tables, like 8x2, 5x5, 2x4, ect.
Example data:
$`0`
$`0`$name
[1] "Vanilla Cream Ale"
$`0`$url
[1] "/homebrew/recipe/view/1633/vanilla-cream-ale"
$`0`$method
[1] "All Grain"
$`0`$style
[1] "Cream Ale"
$`0`$batch
[1] 21.8
$`0`$og
[1] 1.055
$`0`$fg
[1] 1.013
$`0`$abv
[1] 5.48
$`0`$ibu
[1] 19.44
$`0`$color
[1] 4.83
$`0`$`ph mash`
[1] -1
$`0`$fermentables
[,1] [,2] [,3] [,4] [,5]
[1,] "2.381" "American - Pale 2-Row" "37" "1.8" "44.7"
[2,] "0.907" "American - White Wheat" "40" "2.8" "17"
[3,] "0.907" "American - Pale 6-Row" "35" "1.8" "17"
[4,] "0.227" "Flaked Corn" "40" "0.5" "4.3"
[5,] "0.227" "American - Caramel / Crystal 20L" "35" "20" "4.3"
[6,] "0.227" "American - Carapils (Dextrine Malt)" "33" "1.8" "4.3"
[7,] "0.113" "Flaked Barley" "32" "2.2" "2.1"
[8,] "0.34" "Honey" "42" "2" "6.4"
$`0`$hops
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8]
[1,] "14" "Cascade" "Pellet" "6.2" "Boil" "60 min" "11.42" "33.3"
[2,] "14" "Cascade" "Pellet" "6.2" "Boil" "20 min" "6.92" "33.3"
[3,] "14" "saaz" "Pellet" "3" "Boil" "5 min" "1.1" "33.3"
$`0`$`hops Summary`
[,1] [,2] [,3] [,4]
[1,] "28" "Cascade (Pellet)" "18.34" "66.6"
[2,] "14" "saaz (Pellet)" "1.1" "33.3"
$`0`$other
[,1] [,2] [,3] [,4] [,5]
[1,] "2 oz" "pure vanilla extract" "Flavor" "Boil" "0 min."
[2,] "1 oz" "pure vanilla extract" "Flavor" "Bottling" "0 min."
[3,] "1 tsp" "yeast nutrient" "Other" "Boil" "15 min."
[4,] "1 each" "whirlfloc" "Fining" "Boil" "15 min."
[5,] "4 each" "Vanilla beans - in 2oz Vodka" "Other" "Secondary" "0 min."
$`0`$yeast
[1] "Wyeast - Kölsch 2565" "76%" "Low"
[4] "56" "70" "Yes"
$`0`$rating
[1] 0
$`0`$`num rating`
[1] 16
$`0`$views
[1] 289454
As you can see, everything but the "fermentables", "hops", "other", and "yeast" is easy to deal with. I have NO idea what to do with those tables though. I've been scouring StackOverflow and trying different methods, but most of them require the dataset being converted to a dataframe, but I'm getting 'blocked' by these tables. I imagine I would want to isolate them and try to turn it into long data, but I'm not sure how to isolate those. Any suggestions or libraries and documentation I can read up on? Thanks for any help! I've tried to deal with in python previous and still couldn't figure it out.
Edit: I know that I can access them individually via "beer[["0"]][["fermentables"]]", however I can't figure out how to access more than one at a time, which is also throwing me off.
dput() of data for the first beer:
list(`0` = list(name = "Vanilla Cream Ale", url = "/homebrew/recipe/view/1633/vanilla-cream-ale",
method = "All Grain", style = "Cream Ale", batch = 21.8,
og = 1.055, fg = 1.013, abv = 5.48, ibu = 19.44, color = 4.83,
`ph mash` = -1L, fermentables = structure(c("2.381", "0.907",
"0.907", "0.227", "0.227", "0.227", "0.113", "0.34", "American - Pale 2-Row",
"American - White Wheat", "American - Pale 6-Row", "Flaked Corn",
"American - Caramel / Crystal 20L", "American - Carapils (Dextrine Malt)",
"Flaked Barley", "Honey", "37", "40", "35", "40", "35", "33",
"32", "42", "1.8", "2.8", "1.8", "0.5", "20", "1.8", "2.2",
"2", "44.7", "17", "17", "4.3", "4.3", "4.3", "2.1", "6.4"
), .Dim = c(8L, 5L)), hops = structure(c("14", "14", "14",
"Cascade", "Cascade", "saaz", "Pellet", "Pellet", "Pellet",
"6.2", "6.2", "3", "Boil", "Boil", "Boil", "60 min", "20 min",
"5 min", "11.42", "6.92", "1.1", "33.3", "33.3", "33.3"), .Dim = c(3L,
8L)), `hops Summary` = structure(c("28", "14", "Cascade (Pellet)",
"saaz (Pellet)", "18.34", "1.1", "66.6", "33.3"), .Dim = c(2L,
4L)), other = structure(c("2 oz", "1 oz", "1 tsp", "1 each",
"4 each", "pure vanilla extract", "pure vanilla extract",
"yeast nutrient", "whirlfloc", "Vanilla beans - in 2oz Vodka",
"Flavor", "Flavor", "Other", "Fining", "Other", "Boil", "Bottling",
"Boil", "Boil", "Secondary", "0 min.", "0 min.", "15 min.",
"15 min.", "0 min."), .Dim = c(5L, 5L)), yeast = c("Wyeast - Kölsch 2565",
"76%", "Low", "56", "70", "Yes"), rating = 0L, `num rating` = 16L,
views = 289454L)