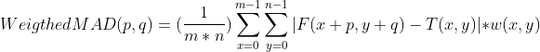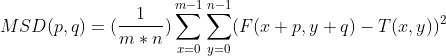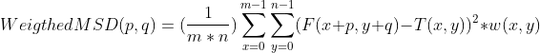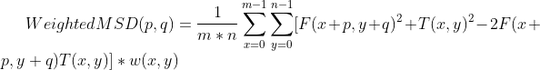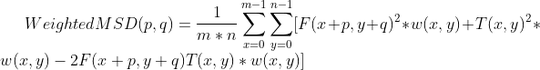I am trying to implement some sort of correlation tracking between a template image and frames of video stream in Python-OpenCV
I am trying to use weighted mean absolute deviation (weighted MAD) as a similarity measure between the template and video frames (object should be at location of minimum MAD.).
the equation I need to do is:
where F is the image, T is the template and w is weight window (same size as template)
I am aware that open-cv provides function which does template matching (i.e.: cv2.matchTemplate). The most close one to MAD is TM_SQDIFF_NORMED which is mean square deviation (MSD), I believe that open-cv implements this equation
which will give the measure of similarity I want if there is a way to implement weight function inside it like this
My question is how can I implement any of weighted MAD or weighted MSD in Open-CV without implementing loops myself (so as not to lose speed) utilizing cv2.matchTemplate function (or similar approach