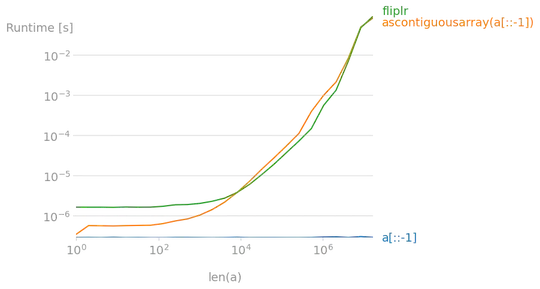
Expanding on what others have said I will give a short example.
If you have a 1D array ...
>>> import numpy as np
>>> x = np.arange(4) # array([0, 1, 2, 3])
>>> x[::-1] # returns a view
Out[1]:
array([3, 2, 1, 0])
But if you are working with a 2D array ...
>>> x = np.arange(10).reshape(2, 5)
>>> x
Out[2]:
array([[0, 1, 2, 3, 4],
[5, 6, 7, 8, 9]])
>>> x[::-1] # returns a view:
Out[3]: array([[5, 6, 7, 8, 9],
[0, 1, 2, 3, 4]])
This does not actually reverse the Matrix.
Should use np.flip to actually reverse the elements
>>> np.flip(x)
Out[4]: array([[9, 8, 7, 6, 5],
[4, 3, 2, 1, 0]])
If you want to print the elements of a matrix one-by-one use flat along with flip
>>> for el in np.flip(x).flat:
>>> print(el, end = ' ')
9 8 7 6 5 4 3 2 1 0