Edited
I have a large table, which starts like this
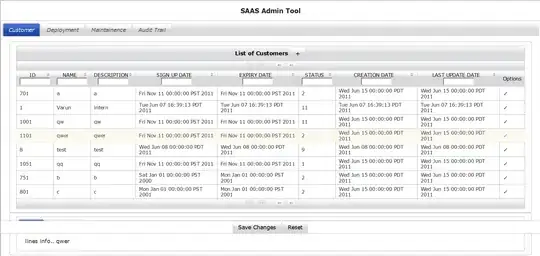
Essentially, it's a table with multiple samples ("samp_id") showing the number ("least") of "taxon" present in each.
I want to transpose/pivot the table to look like this;
i.e. with "taxon" as the top row, with each of the 90 samples in "data" following as a row based on the "least" column, re-named with its "samp_id". So you see what each sample is, as well the value in "least" for each sample in the different "taxon" (which may not be identical across the 90 samples).
Previously, I have separated the data into multiple tibbles based on "samp_id", selected "taxon" and "least", re-named "least" with the "samp_id" then combined the individual tibbles based on "taxon" with full_join using something like the code below, then transposing the combined table
ACLOD_11 = data %>%
filter(samp_id == "ACLOD_11") %>%
select(taxon, least) %>%
rename("ACLOD_11" = least)
ACLOD_12 = data ... #as above, but different samp_id
data_final = list(ACLOD_11, ACLOD_12, ...) %>%
reduce(full_join, by = "taxon")
As I have more data tables to follow after this one with 90 samples, so I want to be able to do this without having to individually separate the data into 100s of tibbles and manually inputting the "samp_id" before joining.
I have currently split the data into 90 separate tibbles based on "samp_id" (there are 90 samples in "data")
data_split = data %>%
group_split(samp_id)
but am unsure if this is the best way to do this, or what I should to next?