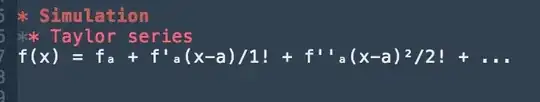
The dput(Q_Sheet) is below. How can properly introduce a second y-axis that is different in scale from the primary axis?
structure(list(Amino_acids = c(4, 12, 20, 28, 32), Protein_length_Ang = c(7,
24, 40, 56, 64), length_no_ratio = c(1.75, 2, 2, 2, 2), Corrected_aa = c(1.24459201924769e-12,
3.71007650662474e-09, 1.10594599229843e-05, 0.0319159404863842,
0.642857142857143), aa_frequency = c(3.99735380592756, 6.96840672963299,
4.58228895300999, 3.12310921028256, 4.67560843680985), T_degC = c(50.3857804818545,
52.8464583426248, 60.0760389538482, 58.1895053328481, 67.628202708438
)), row.names = c(NA, -5L), class = c("tbl_df", "tbl", "data.frame"
), na.action = structure(c(`2` = 2L, `4` = 4L, `6` = 6L), class = "omit"))
`
ggplot(data = Q_Sheet, aes(x = T_degC))+
geom_line(aes(y = Amino_acids), color="red")+
geom_line(aes(y = Corrected_aa), color = "blue") +
scale_y_continuous(name = "Amino_acids", sec.axis = sec_axis(~.*10, name = "Corrected_aa"))
The output is as follows:
<ScaleContinuousPosition>
Range:
Limits: 0 -- 1