One way to do it is to fall back on numpy by first getting the underlying arrays and then looping:
import pandas as pd
from timeit import default_timer as timer
code = 145896
# real df is way bigger
df = pd.DataFrame(data={
'code1': [145896, 800175, 633974, 774521, 416109],
'code2': [100, 800, 600, 700, 400],
'code3': [1, 8, 6, 7, 4]}
)
start = timer()
code1 = df['code1'].values
code2 = df['code2'].values
code3 = df['code3'].values
for _ in range(100000):
desired = code1 == code
desired_code2 = code2[desired][0]
desired_code3 = code3[desired][0]
print(timer() - start) # 0.26 (sec)
The following code adapted from ALollz's answer illustrates the difference in performance of the different techniques with several more added and an increased dataset size range that shows changing behavior affected by orders of magnitude.
import perfplot
import pandas as pd
import numpy as np
def DataFrame_Slice(df, code=0):
return df.loc[df['code1'] == code, 'code2'].iloc[0]
def DataFrame_Slice_Item(df, code=0):
return df.loc[df['code1'] == code, 'code2'].item()
def Series_Slice_Item(df, code=0):
return df['code2'][df['code1'] == code].item()
def with_numpy(df, code=0):
return df['code2'].to_numpy()[df['code1'].to_numpy() == code].item()
def with_numpy_values(code1, code2, code=0):
desired = code1 == code
desired_code2 = code2[desired][0]
return desired_code2
def DataFrameIndex(df, code=0):
return df.loc[code].code2
def with_numpy_argmax(code1, code2, code=0):
return code2[np.argmax(code1==code)]
def numpy_search_sorted(code1, code2, sorter, code=0):
return code2[sorter[np.searchsorted(code1, code, sorter=sorter)]]
def python_dict(code1_dict, code2, code=0):
return code2[code1_dict[code]]
def shuffled_array(N):
a = np.arange(0, N)
np.random.shuffle(a)
return a
def setup(N):
print(f'setup {N}')
df = pd.DataFrame({'code1': shuffled_array(N),
'code2': shuffled_array(N),
'code3': shuffled_array(N)})
code = df.iloc[min(len(df)//2, len(df)//2 + 20)].code1
sorted_index = df['code1'].values.argsort()
code1 = df['code1'].values
code2 = df['code2'].values
code1_dict = {code1[i]: i for i in range(len(code1))}
return df, df.set_index('code1'), code1, code2, sorted_index, code1_dict, code
for relative_to in [5, 6, 7]:
perfplot.show(
setup=setup,
kernels=[
lambda df, _, __, ___, sorted_index, ____, code: DataFrame_Slice(df, code),
lambda df, _, __, ___, sorted_index, ____, code: DataFrame_Slice_Item(df, code),
lambda df, _, __, ___, sorted_index, ____, code: Series_Slice_Item(df, code),
lambda df, _, __, ___, sorted_index, ____, code: with_numpy(df, code),
lambda _, __, code1, code2, sorted_index, ____, code: with_numpy_values(code1, code2, code),
lambda _, __, code1, code2, sorted_index, ____, code: with_numpy_argmax(code1, code2, code),
lambda _, df, __, ___, sorted_index, ____, code: DataFrameIndex(df, code),
lambda _, __, code1, code2, search_index, ____, code: numpy_search_sorted(code1, code2, search_index, code),
lambda _, __, ___, code2, _____, code1_dict, code: python_dict(code1_dict, code2, code)
],
logy=True,
labels=['DataFrame_Slice', 'DataFrame_Slice_Item', 'Series_Slice_Item', 'with_numpy', 'with_numpy_values', 'with_numpy_argmax', 'DataFrameIndex', 'numpy_search_sorted', 'python_dict'],
n_range=[2 ** k for k in range(1, 25)],
equality_check=np.allclose,
relative_to=relative_to,
xlabel='len(df)')
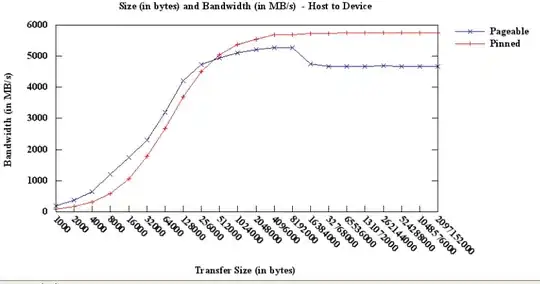
Conclusion: Numpy search sorted is the best performing technique except for very small data sets below 100 mark. Sequential search using the underlying numpy arrays is the next best option for datasets roughly below 100,000 after which the best option is to use the DataFrame index. However, when hitting the multi-million records mark, DataFrame indexes no longer perform well, probably due to hashing collisions.
[EDIT 03/24/2022]
Using a Python dictionary beats all other techniques by at least one order of magnitude.

Note: We are assuming repeated searches within the DataFrame, so as to offset the cost of acquiring the underlying numpy arrays, indexing the DataFrame or sorting the numpy array.