I have chromatograph data (signal) in a pandas df and in one of the signal processing step is to perform peak sharpening as shown in fig below
The reference literature is as follows:
Literature :Paper 1(Peak sharping section ) Paper 2
Algorithm in Literature
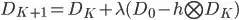
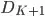 : Deconvolved high resolution data after K + 1 iterations
: Deconvolved high resolution data after K + 1 iterations
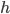 : point spread function
: point spread function
Adaptive point spread function estimation: Peaks are detected in the regularized trace and called as bases with standard classification methods. The called peaks are used to adaptively estimate the local point spread function h. The time-localization parameter d is estimated according to the peak spacing in the segment.
How I can find h for signal data?
Already gone through following
PSF (point spread function) for an image (2D)
How do you extract a point spread function from a fits image?
in all of these it was image data
Sample data
array([[ 31, 49, 1, 44],
[ 36, 48, 0, 47],
[ 43, 47, 0, 53],
[ 50, 44, 0, 63],
[ 59, 41, 0, 75],
[ 68, 40, 1, 90],
[ 78, 40, 6, 107],
[ 87, 41, 12, 123],
[ 99, 43, 20, 140],
[110, 45, 31, 155],
[121, 47, 42, 170],
[131, 48, 53, 182],
[140, 49, 63, 191],
[148, 50, 72, 196],
[155, 51, 79, 196],
[161, 53, 83, 189],
[166, 55, 83, 177],
[169, 58, 80, 160],
[170, 62, 72, 140],
[167, 65, 62, 119],
[161, 70, 51, 100],
[154, 75, 40, 84],
[144, 80, 30, 72],
[132, 86, 23, 65],
[121, 92, 19, 61],
[111, 98, 19, 61],
[106, 102, 23, 63],
[105, 104, 29, 67],
[111, 104, 38, 71],
[123, 102, 48, 75],
[141, 98, 59, 78],
[160, 92, 71, 79],
[179, 85, 85, 78],
[195, 77, 101, 74],
[205, 68, 117, 68],
[208, 59, 133, 61],
[203, 51, 145, 52],
[191, 43, 152, 43],
[173, 37, 154, 35],
[150, 32, 151, 28],
[123, 30, 142, 23],
[ 94, 32, 129, 20],
[ 65, 40, 114, 21],
[ 40, 52, 96, 25],
[ 21, 70, 77, 35],
[ 9, 91, 58, 51],
[ 1, 113, 39, 71],
[ 0, 134, 24, 97],
[ 0, 152, 13, 126],
[ 0, 168, 5, 157],
[ 0, 181, 0, 188],
[ 0, 193, 0, 216],
[ 0, 203, 0, 241],
[ 0, 211, 0, 258],
[ 0, 215, 0, 265],
[ 0, 213, 0, 262],
[ 0, 207, 0, 249],
[ 0, 195, 0, 227],
[ 0, 180, 0, 200],
[ 0, 164, 0, 170],
[ 0, 148, 0, 140],
[ 0, 132, 0, 113],
[ 0, 116, 0, 90],
[ 0, 100, 5, 74],
[ 0, 83, 18, 62],
[ 0, 66, 38, 57],
[ 0, 49, 64, 58],
[ 0, 36, 98, 64],
[ 0, 25, 133, 76],
[ 0, 18, 164, 94],
[ 0, 15, 187, 116],
[ 0, 16, 199, 143],
[ 0, 18, 199, 169],
[ 0, 22, 186, 193],
[ 1, 26, 164, 211],
[ 7, 28, 134, 222],
[ 17, 29, 102, 224],
[ 31, 28, 71, 218],
[ 50, 26, 44, 204],
[ 71, 22, 24, 184],
[ 91, 18, 11, 160],
[106, 13, 4, 134],
[117, 8, 3, 109],
[122, 5, 5, 85],
[120, 2, 10, 64],
[113, 0, 16, 46],
[101, 0, 22, 32],
[ 86, 0, 28, 22],
[ 69, 0, 34, 16],
[ 52, 3, 39, 13],
[ 36, 10, 44, 13],
[ 24, 22, 47, 18],
[ 18, 37, 48, 25],
[ 20, 56, 46, 36],
[ 31, 76, 39, 49],
[ 51, 94, 28, 63],
[ 81, 110, 19, 75],
[118, 123, 10, 85],
[158, 132, 4, 90],
[199, 136, 0, 89],
[236, 135, 0, 84],
[265, 131, 0, 73],
[282, 122, 0, 59],
[286, 110, 0, 44],
[277, 95, 0, 29],
[256, 79, 0, 17],
[226, 61, 0, 8],
[189, 44, 0, 2],
[150, 29, 6, 0],
[112, 17, 19, 0],
[ 77, 8, 41, 0],
[ 49, 3, 74, 0],
[ 28, 0, 117, 0],
[ 15, 0, 168, 3],
[ 7, 0, 224, 12],
[ 5, 0, 280, 28],
[ 5, 0, 333, 53],
[ 5, 0, 379, 87],
[ 5, 0, 411, 130],
[ 4, 0, 425, 178],
[ 2, 0, 419, 226],
[ 1, 0, 393, 271],
[ 0, 0, 350, 307],
[ 0, 0, 299, 329],
[ 0, 0, 248, 334],
[ 0, 0, 206, 320],
[ 0, 0, 178, 289],
[ 0, 0, 167, 246],
[ 0, 0, 173, 196],
[ 0, 0, 192, 146],
[ 0, 0, 217, 100],
[ 0, 0, 246, 61],
[ 0, 0, 275, 33],
[ 0, 0, 301, 15],
[ 0, 0, 326, 4],
[ 0, 0, 351, 0],
[ 0, 0, 377, 0],
[ 0, 0, 403, 0],
[ 0, 0, 430, 0],
[ 0, 0, 456, 0],
[ 0, 0, 484, 0],
[ 0, 0, 510, 0],
[ 0, 3, 535, 0],
[ 0, 7, 555, 0],
[ 0, 14, 569, 0],
[ 0, 22, 574, 0],
[ 0, 33, 572, 0],
[ 0, 44, 565, 0],
[ 0, 55, 555, 0],
[ 0, 66, 548, 0],
[ 1, 76, 546, 0],
[ 9, 84, 550, 0],
[ 27, 91, 555, 0],
[ 53, 94, 558, 0],
[ 88, 95, 554, 0],
[132, 94, 536, 2],
[178, 91, 503, 7],
[219, 86, 453, 15],
[252, 82, 390, 24],
[273, 77, 317, 36],
[279, 72, 240, 44],
[270, 69, 165, 49],
[247, 69, 102, 48],
[214, 73, 55, 41],
[173, 83, 23, 30],
[132, 98, 5, 19],
[ 93, 121, 0, 10],
[ 59, 150, 0, 3],
[ 34, 186, 0, 0],
[ 17, 227, 0, 0],
[ 6, 271, 0, 0],
[ 1, 315, 0, 0],
[ 0, 354, 0, 0],
[ 0, 384, 0, 5],
[ 0, 404, 0, 13],
[ 0, 413, 0, 25],
[ 0, 411, 0, 39],
[ 0, 398, 0, 55],
[ 0, 378, 0, 67],
[ 0, 350, 0, 75],
[ 0, 315, 0, 76],
[ 0, 276, 9, 72],
[ 0, 235, 28, 64],
[ 0, 198, 60, 53],
[ 2, 164, 107, 42],
[ 13, 139, 172, 30],
[ 36, 120, 242, 20],
[ 78, 109, 310, 12],
[143, 103, 367, 7],
[233, 101, 404, 3],
[344, 101, 414, 1],
[467, 103, 394, 0],
[591, 103, 350, 0],
[702, 100, 287, 0],
[786, 94, 215, 0],
[831, 84, 144, 0],
[833, 70, 86, 0],
[792, 54, 44, 4],
[713, 38, 17, 16],
[606, 24, 4, 36]], dtype=int64)