I have a vector of data, which contains integers in the range -20 20.
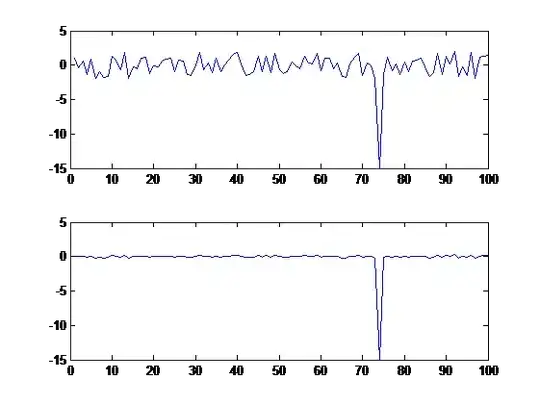
Bellow is a plot with the values:
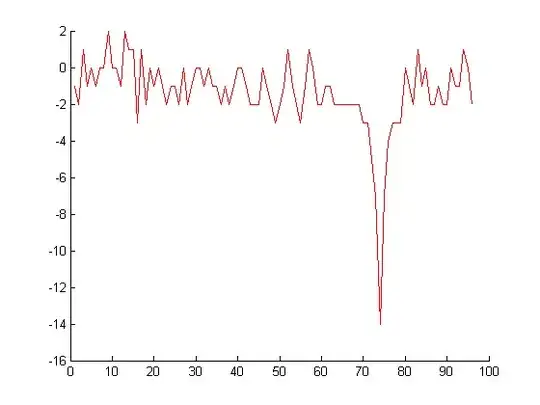
This is a sample of 96 elements from the vector data. The majority of the elements are situated in the interval -2, 2, as can be seen from the above plot.
I want to eliminate the noise from the data. I want to eliminate the low amplitude peaks, and keep the high amplitude peak, namely, peaks like the one at index 74.
Basically, I just want to increase the contrast between the high amplitude peaks and low amplitude peaks, and if it would be possible to eliminate the low amplitude peaks.
Could you please suggest me a way of doing this?
I have tried mapstd function, but the problem is that it also normalizes that high amplitude peak.
I was thinking at using the wavelet transform toolbox, but I don't know exact how to reconstruct the data from the wavelet decomposition coefficients.
Can you recommend me a way of doing this?